Analysis
Misc
- Also see
- Domain Knowledge >> Epidemiology >> Disease Mapping
- Mathematics, Statistics >> Multivariate >> Depth
- Outlier detection and robust mean calculation for multivariate geospatial and spatio-temperal data
- Packages
- {exactextractr} - Written in C++ to quickly and accurately summarize raster values over polygonal areas, commonly referred to as zonal statistics
- Has the traditional descriptive statistics and more
- {lazysf} (README has better documentation)- Provides a {dplyr} backend for any vector data source (aka shapefiles) readable by GDAL (e.g. gpkg, shp, geojson).
- Uses
gdalraster::GDALVectorto talk to GDAL and {dbplyr} for SQL translation, giving you lazy evaluation of spatial data through familiar {dplyr} verbs. - For larger datasets, enable GDAL’s columnar Arrow C stream interface for faster data transfer (link)
- Uses
- {sfhotspot} (Thread, Intro)- Functions to identify and understand clusters of points (typically representing the locations of places or events)
- Built for novice users, by choosing reasonable default values and doing as much as possible for users behind the scenes. There’s also a vignette to take more control for more experienced users.
- {spopt}, {spopt} - Spatial Optimization
- Regionalization, facility location, and transportation-oriented modeling
- Build contiguous regions from your data with algorithms like Max-P, SKATER, and AZP. Useful for political redistricting, sales territory delineation, and more
- Solve facility location problems using classic algorithms like P-Median and MCLP, and consider more advanced scenarios like facility capacity and real estate costs
- Analyze retail market share and cannibalization with the classic Huff model
- Which store a customer is likely to visit?
- Sales potential per location?
- How new stores reshape the competitive landscape?
- Use built-in Euclidean distance for speed, or bring your own travel-time cost matrices from packages like r5r for more realistic modeling.
- {exactextractr} - Written in C++ to quickly and accurately summarize raster values over polygonal areas, commonly referred to as zonal statistics
- Boundary Analysis
- The assessment of whether significant geographic boundaries are present and whether the boundaries of multiple variables are spatially correlated.
- Notes from BoundaryStats: An R package to calculate boundary overlap statistics
- Packages
- {BoundaryStats} - Functions for boundary and boundary overlap statistics
- Boundaries are areas in which spatially distributed variables (e.g., bird plumage coloration, disease prevalence, annual rainfall) rapidly change over a narrow space.
- Boundary Statistics
- The length of the longest boundary
- The number of cohesive boundaries on the landscape
- Boundary Overlap Statistics
- The amount of direct overlap between boundaries in variables \(A\) and \(B\)
- The mean minimum distance between boundaries in \(A\) and \(B\) (i.e. minimums are measured within \(A\))
- For instance, the minimum distance between \(\text{boundary}_{A,i}\) and \(\text{boundary}_{A,j}\)
- The mean minimum distance from boundaries in \(A\) to boundaries in \(B\)
- Use Cases
- By identifying significant cohesive boundaries, researchers can delineate relevant geographic sampling units (e.g., populations as conservation units for a species or human communities with increased disease risk).
- Associations between the spatial boundaries of two variables can be useful in assessing the extent to which an underlying landscape variable drives the spatial distribution of a dependent variable.
- Identifying neighborhood effects on public health outcomes, including COVID-19 infection risk or spatial relationships between high pollutant density and increased disease risk.
Terms
- Areal (aka Lattice) Data - Data in a study region that is partitioned into a limited number of areas, with outcomes being aggregated or summarized within those areas (instead of at points). Examples include:
- Population density
- Disease rates
- Income levels
- Crime statistics
- Educational attainment
- Unemployment rates
- Block Support - When the variable value summarizes all points in the geometry. (Also see Point Support)
- An aggregate; the feature is a single observation with a value that is associated with the entire geometry
- Examples
- Population, either as number of persons or as population density
- Other socio-economic variables, summarised by area
- Average reflectance over a remote sensing pixel
- Total emission of pollutants by region
- Bblock mean NO concentrations, such as obtained by block kriging over square blocks or by a dispersion model that predicts areal mean
- Buffer - a zone around a geographic feature containing locations that are within a specified distance of that feature, the buffer zone. A buffer is likely the most commonly used tool within the proximity analysis methods. Buffers are usually used to delineate protected zones around features or to show areas of influence.
- Catchment - The area inside any given polygon is closer to that polygon’s point than any other. Refers to the area of influence from which a retail location, such as a shopping center, or service, such as a hospital, is likely to draw its customers. (also see Retail >> Catchment)
- Data Fusion - The primary idea is to combine different types of spatial data. This can include:
- Remote Sensing Data: Satellite imagery (multispectral, hyperspectral, SAR, LiDAR), drone data, aerial photographs. These often provide wide coverage and different spectral/spatial resolutions.
- In-situ Data: Field measurements, sensor networks, ground surveys. These offer high accuracy at specific locations.
- GIS Databases: Vector data (points, lines, polygons), land use maps, elevation models.
- Socio-economic Data: Population density, demographic information.
- Temporal Data: Combining data from the same source over different time periods (e.g., monitoring deforestation over years).
- Extensive Variables - Correspond to amounts, associated with a physical size (length, area, volume, counts of items). (Also see Intensive Variables) (source)
- Example: Population Count
- It is associated with an area, and if that area is cut into smaller areas, the population count needs to be split too.
- Because population is rarely uniform over space, this does not need to be done proportionally to the smaller areas, but the sum of the population count for the smaller areas needs to equal that of the total.
- Example: Population Count
- Feature Attributes - The properties of features (“things”) that do not describe the feature’s geometry. (Not sure I get the difference between describes and derived in this context)
- Properties Derived From Geometry:
- Length of a LINESTRING
- Area of a POLYGON
- Properties Not Derived From Geometry:
- Name of a street or county
- Number of people living in a country
- Type of a road
- Soil type in a polygon from a soil map
- Opening hours of a shop
- Body weight or heart beat rate of an animal
- NO2 concentration measured at an air quality monitoring station
- Time Properties:
- Date of birth of a person
- Construction year of a road
- Air Quality (function of both space and time)
- Properties Derived From Geometry:
- Identitiy Variable - When the associated geometry uniquely identifies the variable’s value (there are no other geometries with the same value)
- Example: An arbitrary point (or region) inside a county is still part of the county and must have the same value for county name, but it no longer identifies the (entire) geometry corresponding to that county
- Intensive Variables - Variables that do NOT have values proportional to support: if the area is split, values may vary but on average remain the same. (Also see Extensive Variables)(source)
- Example: Population Density
- If an area is split into smaller areas, population density is not split similarly: the sum of population densities for the smaller areas is a meaningless measure, as opposed to the average of the population densities which will be similar to the density of the total area.
- Example: Population Density
- Point Support - When the variable value applies to every point (Also see Block Support)
- The attribute value is valid everywhere in or over the geometry; we can think of the feature as consisting of an infinite number of points that all have this attribute value
- Examples
- Land use for a land use polygon
- Rock units or geologic strata in a geological map
- Soil type in a soil map
- Elevation class in an elevation map that shows elevation as classes
- Climate zone in a climate zone map
- Realization - It’s one specific, observed outcome or dataset from a spatial random process, i.e. random sample.
- Since it is impossible to sample or measure every point in a spatial domain, the spatial data we collect represents just one instance of a larger, underlying probability model.
- Spatial modeling techniques, such as geostatistical simulation, use this observed realization to predict and quantify uncertainty about the spatial variable at unobserved locations.
- Support - The spatial area an attribute value refers to
- Spatial Functional Data - Data comprising curves or functions that are recorded at each spatial location.
- Trans-Gaussian Variables - Variables whose distribution can be transformed into a normal distribution, e.g. lognormal variables
Descriptive
Average elevation for areas (source)
pacman::p_load( sf, dplyr, purrr, geo ) # Read the CRS of the source elevation_crs <- st_layers("data/ns-water_elevation.fgb")$crs[[1]] # Transform the bounds to the CRS of the source bounds_poly <- bounds_lonlat |> st_transform(elevation_crs) |> st_as_sfc() # Read just the data you need elevation <- st_read( "data/ns-water_elevation.fgb", wkt_filter = st_as_text(bounds_poly) ) lakes <- read_sf("data/ns-water_water-poly.fgb") # filter lakes and convert crs lakes <- lakes |> filter(st_intersects(lakes, bounds_poly[[1]], sparse = FALSE)) |> st_transform(elevation_crs) ######### methods for calculating mean elevation ######### # 1. # long way # lakes$mean_elev <- lakes$geometry |> # map_dbl(~{ # elev_is_relevant <- st_intersects(elevation, .x, sparse = FALSE) # mean(elevation$ZVALUE[elev_is_relevant]) # }) # 2. lakes |> mutate(row_id = row_number()) |> st_join(elevation, join = st_intersects) |> group_by(row_id) |> summarise(mean_elev = mean(ZVALUE.y)) # 3. index <- geos_strtree(elevation) lakes$geometry |> as_geos_geometry() |> map_dbl(~{ elevation_candidates <- elevation[geos_strtree_query(index, .x)[[1]]] elevations <- elevation_candidates[geos_prepared_intersects(elevation_candidates, .x)] mean(geos_z(elevations)) })- Finds the average elevation for some lakes
- The “long way” takes about about 223 sec
- The sf join way takes about 10 sec
- The geos way takes about 2.5 sec
geos_strtreecreates a spatial index which speeds things up
Proximity Analysis
- Example: Basic Workflow
Data: Labels, Latitude, and Longitude
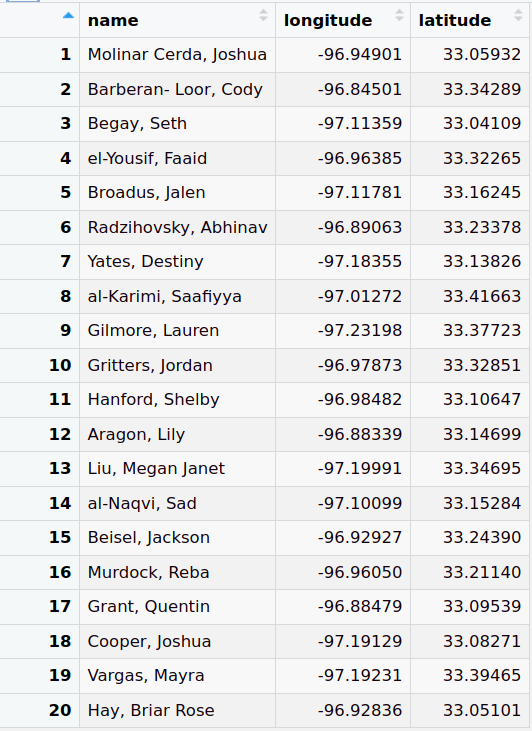
Create Simple Features (sf) Object
customer_sf <- customer_table %>% sf::st_as_sf(coords = c("longitude", "latitude"), crs = 4326)- Merges the longitude and latitude columns into a geometry column and transforms the coordinates in that column according to projection (e.g.
crs = 4326)
- Merges the longitude and latitude columns into a geometry column and transforms the coordinates in that column according to projection (e.g.
View points on a map
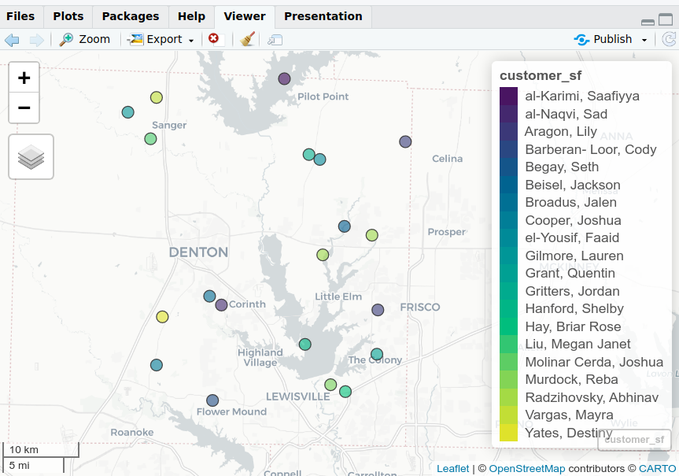
mapview::mapview(customer_sf)Create Buffer Zones
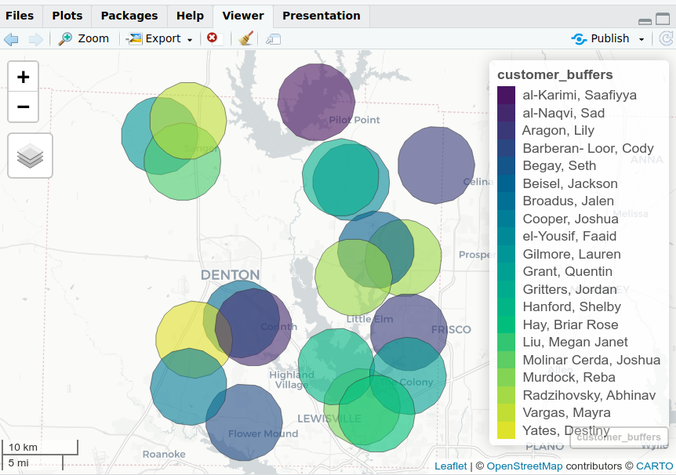
customer_buffers <- customer_sf %>% sf::st_transform(26914) %>% sf::st_buffer(5000) mapview::mapview(customer_buffers)- Most of projections use meters, and based on the size of the circles as related to the size of Denton, TX, I’m guessing the radius of each circle is 5000m. Although, that still looks a little small.
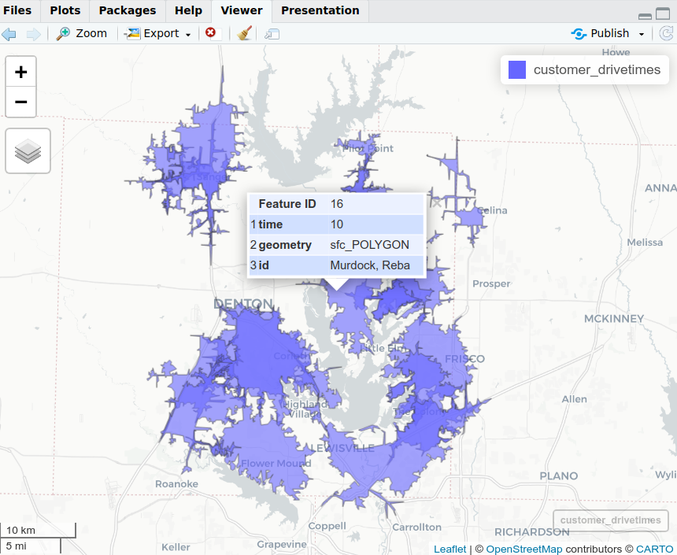
Create Isochrones
customer_drivetimes <- customer_sf %>% mapboxapi::mb_isochrone(time = 10, profile = "driving", id_column = "name") mapview::mapview(customer_drivetimes)- 10 minutes drive-time from each location
- time (minutes): The maximum time supported is 60 minutes. Reflects traffic conditions for the date and time at which the function is called.
- If reproducibility of isochrones is required, supply an argument to the depart_at argument.
- depart_at: Specifying a time makes it a time-aware isochrone. Useful for modeling peak business hours or rush hour traffic, etc.
- e.g. Adding depart_at = “2024-01-27T17:30” to the isochrone above gives you a 10-minute driving isochrone with predicted traffic at 5:30pm tomorrow
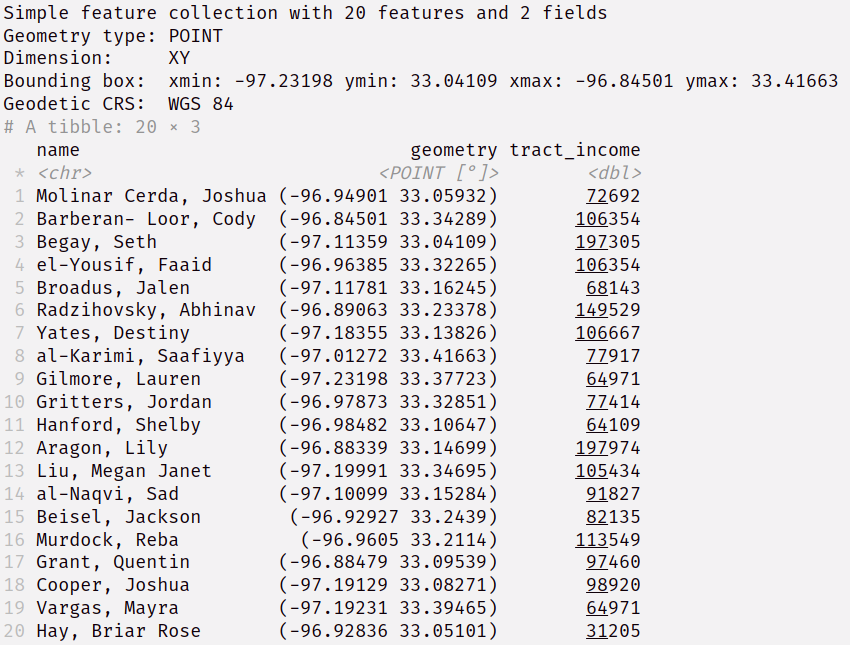
Add Demographic Data
denton_income <- tidycensus::get_acs( geography = "tract", variables = "B19013_001", state = "TX", county = "Denton", geometry = TRUE ) %>% select(tract_income = estimate) %>% sf::st_transform(st_crs(customer_sf)) customers_with_income <- customer_sf %>% sf::st_join(denton_income) customers_with_incomeAdds median income estimate according to the census tract each person lives in.
Joins on the geometry variable
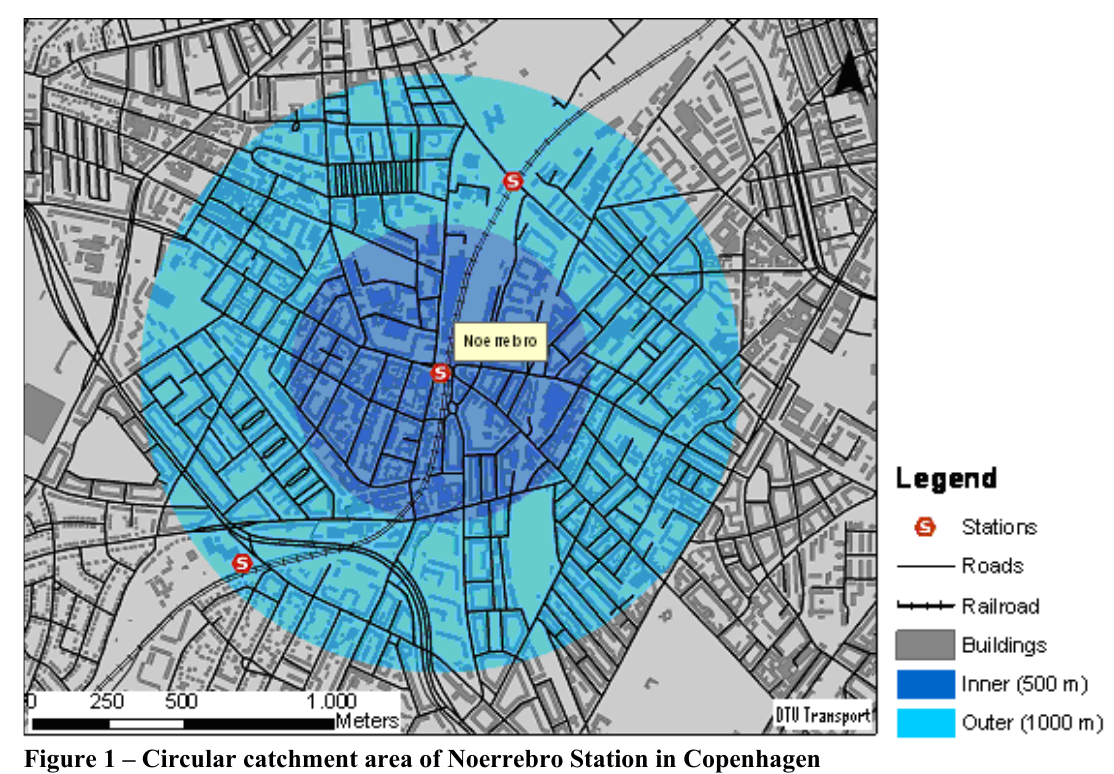
- Circular Buffer Approach
- Notes from GIS-based Approaches to Catchment Area Analyses of Mass Transit
- The simplest and most common used approach to make catchment areas of a location is to consider the Euclidean distance from the location.
- Due to limitations (See below), it’s best suited for overall analyses of catchment areas.
- Often the level of detail in the method has been increased by dividing the catchment area into different rings depending on the distance to the station.
- Limitation: Does not take the geographical surroundings into account.
- Example: In most cases, the actual walking distance to/from a location is longer than the Euclidean distance since there are natural barriers like rivers, buildings, rail tracks etc.
- This limitation is often coped with by applying a detour factor that reduces the buffer distance to compensate for the longer walking distance.
- However, in cases where the length of the detours varies considerably within the location’s surroundings, this solution is not very precise.
- Furthermore, areas that are separated completely from a location, e.g. by rivers, might still be considered as part of the location’s catchment area
- Example: In most cases, the actual walking distance to/from a location is longer than the Euclidean distance since there are natural barriers like rivers, buildings, rail tracks etc.
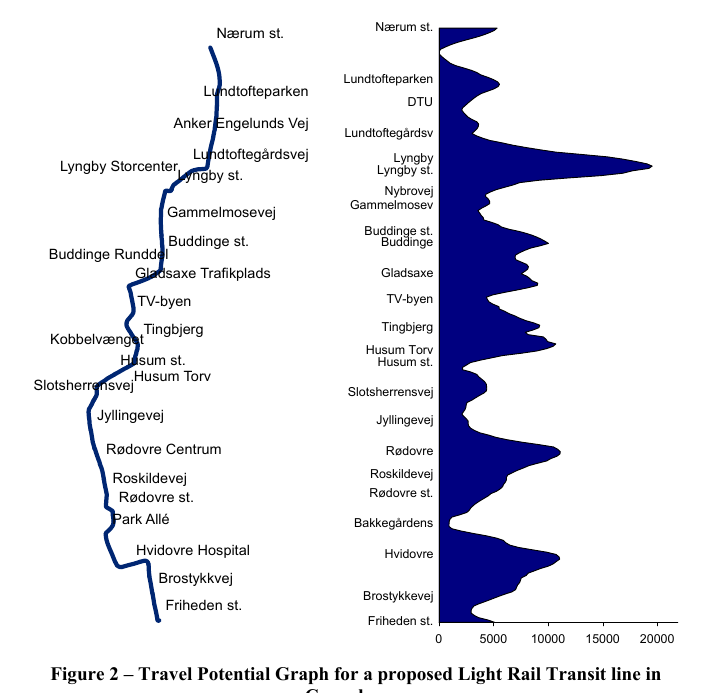
- Use Case: Ascertain Travel Potential to Determine Potential Station Locations
Every 50m along the proposed transit line, calculate the travel potential for that buffer area
- Using the travel demand data for that buffer area, calculate travel potential
Travel Potential Graph
- Left side represents the transit line.
- Right Side
- Y-Axis are locations where buffer areas were created.
- X-Axis: Travel Potential
- Not sure if that is just smoothed line with a point estimate of Travel Potential at each location or how exactly those values are calculated.
- 50m isn’t a large distance so maybe all the locations aren’t shown on the Y-Axis and the number of calculations produces an already, mostly, smooth line on it’s own.
- Partitioning a buffer zone into rings or some kind of interpolation could provided more granular estimates around the central buffer location.
- Not sure if that is just smoothed line with a point estimate of Travel Potential at each location or how exactly those values are calculated.