Mixed Effects
Misc
- Packages
- {Inflongitudinal} - Provides methods for detecting influential subjects in longitudinal data, particularly when observations are collected at irregular time points.
- Identifies subjects whose response trajectories deviate substantially from population-level patterns, helping to diagnose anomalies and undue influence on model estimates.
- {performance} - Utilities for computing indices of model quality and goodness of fit.
- Metrics such as R-squared (R2), root mean squared error (RMSE), intraclass correlation coefficient (ICC), etc.
- Checks for overdispersion, zero-inflation, convergence, or singularity.
- {ggResidpanel} - Standard residual plots with appropriate residual types depending on whether it’s a glm or lm type of model. Uses {ggplot2}
- Has convenience function for plotting residuals vs predictors
- Natively supports models of type “lm”, “glm”, “lme”, “lmer”, “glmer”, and “lmerTest” out of the box
- Can support other models types with
resid_auxpanelif you want to supply your own residuals and fitted/predicted values.
- {ivd} - Fit mixed-effects location scale models with spike-and-slab priors on the location random effects to identify units with unusual residual variances
- {marginme} - Estimation of Relative Risks, Risk Differences, and Marginal Effects from Mixed Models Using Marginal Standardization
- Supports lme4, glmmTMB, or glmmrBase
- {marginaleffects} and {emmeans} handle these packages (I think) and more, but who knows — maybe it’ll come in handy
- {Inflongitudinal} - Provides methods for detecting influential subjects in longitudinal data, particularly when observations are collected at irregular time points.
Warnings and Messages
- boundary (singular) fit: see help(‘isSingular’)
- From {lme4}
- Singular means the estimated variance-covariance matrices are less than full rank i.e. some “dimensions” of the variance-covariance matrix have been estimated as exactly (or nearly) zero
- Also typically leads to random effect correlations of -1 or 1 because this boundary includes the cases where some varying coefficients have zero variance or a varying coefficient is a linear combination of the other varying coefficients.
- Often (30% to 40%) happens when the number of groups is small, data are noisy, and a varying slopes model is fit. (e.g. 5 to 10 groups with n = 10 in each) (Gelman)
- Not a warning, but it’s in red letters. Gelman says not to fret about it. If you have a good fitting model in other respects, then there’s no reason to chuck it because of this message. (Gelman)
- They are statistically well-defined (i.e. sensible Maximum Likelihood estimates)
- For example, it’s better to have the larger standard errors of the varying slopes and intercepts model than the smaller standard errors of a varying intercepts model.
- Problems with a singular model
- May have poor power
- Higher chances of misconvergence
- It may be computationally difficult to compute profile confidence intervals for such models
- Standard inferential procedures such as Wald statistics and likelihood ratio tests may be inappropriate.
help('isSingular')(essentially) gives the following advice (see docs for more details)- Design your experiments better. You should simulate data to find out whether these types of models would result in a singularity.
- Use some form of model selection to choose a model that balances predictive accuracy and overfitting/type I error
- “Keep it maximal”, i.e. fit the most complex model consistent with the experimental design, removing only terms required to allow a non-singular fit.
- Use {blme} (partially Bayesian) which has MAP estimates and a regularizing prior to move covariance matrices away from singularity
- Use {rstanarm} or {brms} (fully bayesian) with informative priors and gives parameters that average over the uncertainty in the random effects parameters.
- Can also use averaged data, which might lose some statistical efficiency but will allow you to use simpler statistical methods (See section 2.3, “Simple paired-comparisons analysis” in Gelman)
Convergence Issues
- See
?lme4::convergence - Gist of Checks (Choe)
- Set a more lenient tolerance for the gradient
- Turn off gradient optimization (?)
- Use stricter precision
- Restart the fit from the reported optimum, or from a point perturbed slightly away from the reported optimum
- Test all optimizers: To assess whether convergence warnings render the results invalid, or to the contrary, the results can be deemed valid in spite of the warnings, Bates et al. (2023) suggest refitting models affected by convergence warnings with a variety of optimizers. (article)
If all optimizers converge to values that are practically equivalent, then consider the convergence warnings to be false positives.
Example: {lme4}
library(lme4) library(dfoptim) library(optimx) fit <- lmer(fatigue ~ spin * reg + (1|ID), data = testdata, REML = TRUE) # Refit model using all available algorithms multi_fit <- allFit(fit) #> bobyqa : [OK] #> Nelder_Mead : [OK] #> nlminbwrap : [OK] #> nmkbw : [OK] #> optimx.L-BFGS-B : [OK] #> nloptwrap.NLOPT_LN_NELDERMEAD : [OK] #> nloptwrap.NLOPT_LN_BOBYQA : [OK] summary(multi_fit)$fixef #> (Intercept) spin reg spin:reg #> bobyqa -2.975678 0.5926561 0.1437204 0.1834016 #> Nelder_Mead -2.975675 0.5926559 0.1437202 0.1834016 #> nlminbwrap -2.975677 0.5926560 0.1437203 0.1834016 #> nmkbw -2.975678 0.5926561 0.1437204 0.1834016 #> optimx.L-BFGS-B -2.975680 0.5926562 0.1437205 0.1834016 #> nloptwrap.NLOPT_LN_NELDERMEAD -2.975666 0.5926552 0.1437196 0.1834017 #> nloptwrap.NLOPT_LN_BOBYQA -2.975678 0.5926561 0.1437204 0.1834016- {lme4::allfit} requires the other two packages to fit all available optimizers
- Article also has a custom plotting function to visually compare the results
Metrics
Also see
- Mixed Effects, BMLR >> Ch.8 >> Model Building Workflow >> Unconditional Means for an ICC example
- Mixed Effects, General >> Examples >> Example 1 >> Varying Intercepts for an ICC example
Restricted Maximum Likelihood (REML)
- It is a form of the likelihood function that accounts for the loss of degrees of freedom due to the estimation of the fixed effects parameters
- The REML criterion is minimized during the model estimation process, and the estimation algorithm iterates until it converges to a point where the REML criterion cannot be further minimized. The value of the REML criterion at this convergence point is reported in the model summary.
- A lower value is better, but the REML criterion value itself is not directly interpretable. it is mainly used for comparing the relative fit of different models to the same data.
- Does not account for model complexity or the principle of parsimony, so it should be used in conjunction with domain knowledge and other metrics.
Check Degrees of Freedom (DOF) in Fixed Effects Estimates
The degrees of freedom for the estimates should be close to the number of subjects (aka units)
Large degrees of freedom in comparison to the number of subjects indicates a misspecification of the model. (e.g. 60 subjects and dofs are close to the number of observations)
- Degrees of Freedom that are less than then number of subjects are fine and could be due to substantially imbalanced numbers of observations among the subjects.
Also papers should report the dofs.
Example
library(lmerTest) data("sleepstudy") lmer_mod <- lmer(Reaction ~ 1 + Days + (1 + Days | Subject), sleepstudy, REML = 0) summary(lmer_mod) #> Fixed effects: #> Estimate Std. Error df t value Pr(>|t|) #> (Intercept) 251.405 6.632 18.001 37.907 < 2e-16 *** #> Days 10.467 1.502 18.000 6.968 1.65e-06 *** lmer_mod2 <- lmer(Reaction ~ 1 + Days + (1 | Subject), sleepstudy, REML = 0) summary(lmer_mod2) #> Fixed effects: #> Estimate Std. Error df t value Pr(>|t|) #> (Intercept) 251.4051 9.5062 24.4905 26.45 <2e-16 *** #> Days 10.4673 0.8017 162.0000 13.06 <2e-16 ***- There are 18 subjects, so the first model is a well-specified model
- The dof spikes to 162 for the Days fixed effect which is close to the number of total observations at 180. This indicates a misspecified model.
- The 24.4905 is actually okay for the intercept.
- Note that you need to used {lmerTest} and not {lme4} to fit the model in order to get the dofs and pvals. Although there’s probably a helper function in {lmerTest} since it loads {lme4} that will give the same results if you fit the model using {lme4}
InterClass Coefficient (ICC): The proportion of variation that is between-cases
\[ \rho = \frac{\sigma_0}{\sigma_0 + \sigma_\epsilon} \]
Where \(\sigma_0\) is the between-case variance and \(\sigma_\epsilon\) is the within-case variance.
1-ICC is the proportion of variation within cases
- Although {sjPlot::tab_model} does compute a ICC for varying slopes models
Statistical power is a function of ICC (article)
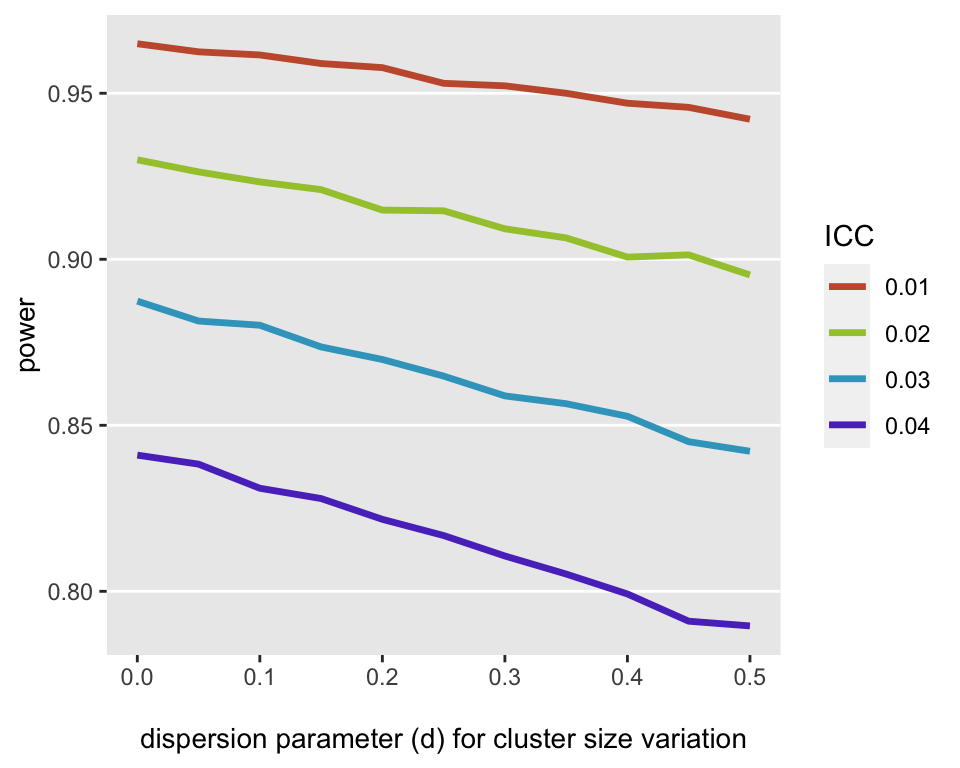
- Both higher ICCs and cluster size variability lead to reduced power
- The dispersion parameter is a parameter used in the data simulation
Guideline: ICC > 0.1 is generally accepted as the minimal threshold for justifying the use of Mixed Effects Model
Example 1: {sjPlot}
library(sjPlot) tab_model(lme_fit)- Might need {lmerTest} loaded to get coefficient pvals
- Also calculates two R2 values
- Marginal: proportion of variance explained by the fixed effects only
- Conditional: proportion of variance explained by the fixed effects and random effects
Example 2: {performance}
data("stroop", package = "afex") stroop_sub <- subset(stroop, !is.na(rt)) stroop_agg <- stroop_sub |> summarize(mean_rt = mean(rt), .by = c("pno", "congruency", "condition")) MLM <- lmerTest::lmer(rt ~ congruency * condition + (1 | pno), data = stroop_agg) performance::icc(MLM) #> # Intraclass Correlation Coefficient #> #> Adjusted ICC: 0.752 #> Unadjusted ICC: 0.632- Adjusted ICC only relates to the random effects
- Unadjusted ICC takes the random effects and fixed effects variances into account, more precisely, the fixed effects variance is added to the denominator of the formula to calculate the ICC
- Don’t pay attention to the aggregation. It was done just to illustrate a point about the necessity of varying slopes even in cases where there is no repeated measure
R2, AICc, BICc
Packages
- {greybox::AICc} or {greybox::BICc} for corrected versions of the information criterion metrics. Supposed to work on any model with
logLikandnobsmethods- Lower is Better (which is the model that minimizes the information loss)
- {glmm.hp} - Hierarchical Partitioning of Marginal R2 for Generalized Mixed-Effect Models
- Calculates individual contributions of each predictor (fixed effects) towards marginal R2
- {greybox::AICc} or {greybox::BICc} for corrected versions of the information criterion metrics. Supposed to work on any model with
Example: From ICC >> Example 2
performance::r2(MLM) #> # R2 for Mixed Models #> #> Conditional R2: 0.792 #> Marginal R2: 0.161 greybox::AICc(MLM) #> [1] -5910.074 greybox::BICc(MLM) #> [1] -5904.154- Nakagawa’s R2 is used
- Conditional R2: Takes both the fixed and random effects into account.
- Marginal R2: Considers only the variance of the fixed effects.
Residuals
Misc
- {DHARMa} - Built for Mixed Effects Models for count distributions but also handles lm, glm (poisson) and MASS::glm.nb (neg.bin)
Binned Residuals
It is not useful to plot the raw residuals, so examine binned residual plots
Misc
- {arm} will mask some {tidyverse} functions, so don’t load whole package
Look for :
- Patterns
- Nonlinear trend may be indication that squared term or log transformation of predictor variable required
- If bins have average residuals with large magnitude
- Look at averages of other predictor variables across bins
- Interaction may be required if large magnitude residuals correspond to certain combinations of predictor variables
Process
- Extract raw residuals
- Include
type.residuals = "response"in thebroom::augmentfunction to get the raw residuals
- Include
- Order observations either by the values of the predicted probabilities (or by numeric predictor variable)
- Use the ordered data to create g bins of approximately equal size.
- Default value: g = sqrt(n)
- Calculate average residual value in each bin
- Plot average residuals vs. average predicted probability (or average predictor value)
- Extract raw residuals
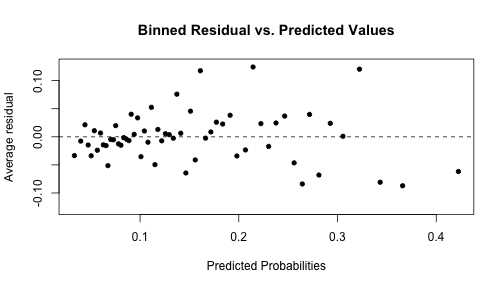
-
arm::binnedplot(x = risk_m_aug$.fitted, y = risk_m_aug$.resid, xlab = "Predicted Probabilities", main = "Binned Residual vs. Predicted Values", col.int = FALSE) -
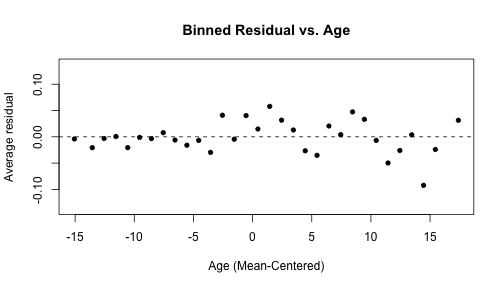
arm::binnedplot(x = risk_m_aug$ageCent, y = risk_m_aug$.resid, col.int = FALSE, xlab = "Age (Mean-Centered)", main = "Binned Residual vs. Age")
Check that residuals have mean zero:
mean(resid(mod))Check that residuals for each level of categorical have mean zero
risk_m_aug %>% group_by(currentSmoker) %>% summarise(mean_resid = mean(.resid))Check for normality.
# Normal Q-Q plot qqnorm(resid(mod)) qqline(resid(mod))Check normality per categorical variable level
Example: 3 levels
## by level par(mfrow=c(1,3)) qqnorm(resid(mod)[1:6]) qqline(resid(mod)[1:6]) qqnorm(resid(mod)[7:12]) qqline(resid(mod)[7:12]) qqnorm(resid(mod)[13:18]) qqline(resid(mod)[13:18])- Data should be sorted by random variable level before modeling. Otherwise you could column bind the residuals to the original data. Then, group by random variable and make q-q plots for each group
Examples
Example 1: {performance}l Varying Intercepts, Varying Slopes
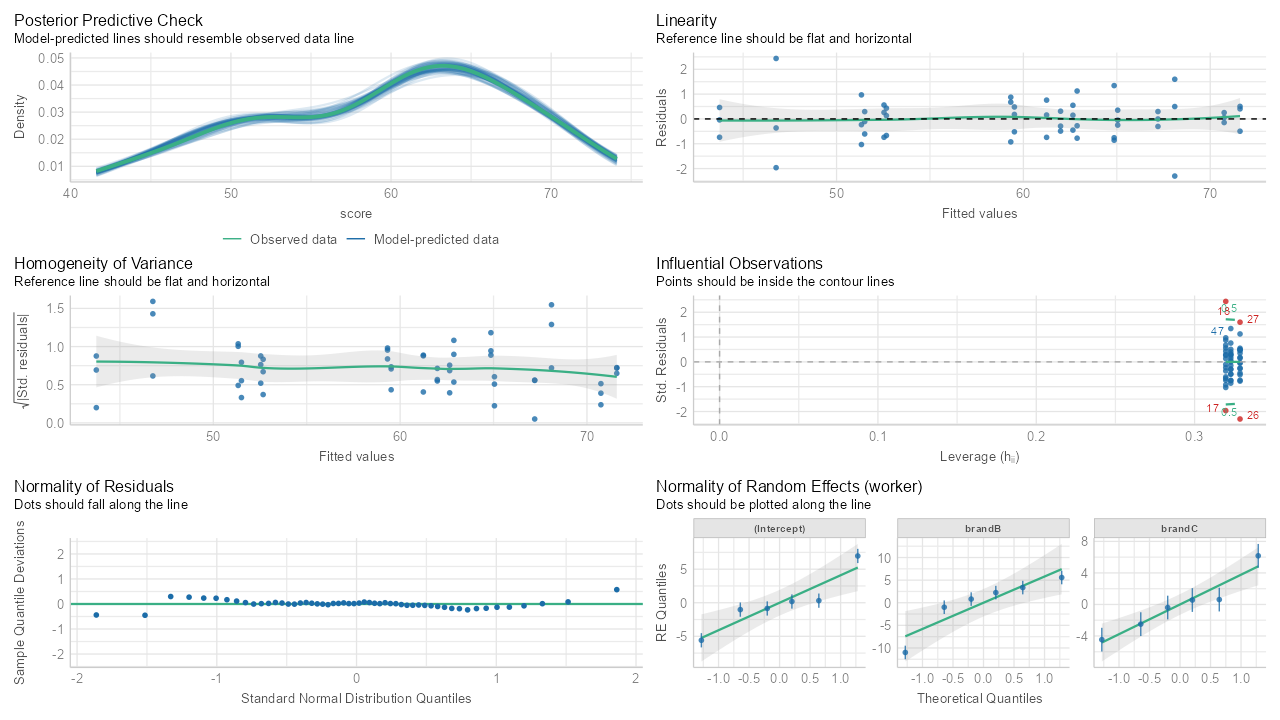
library(performance); library(lmerTest) library(dplyr) data(Machines, package = "nlme") machines <- Machines |> mutate(Worker = factor(Worker, levels = 1:6, ordered = FALSE)) |> rename(worker = Worker, brand = Machine) |> group_by(brand, worker) |> mutate(avg_score = mean(score)) mod_bw_si <- lmer(score ~ brand + (1 + brand | worker), data = machines) check_model(mod_bw_si) model_performance(mod_bw_si) # Indices of model performance #> AIC | AICc | BIC | R2 (cond.) | R2 (marg.) | ICC | RMSE | Sigma #> ----------------------------------------------------------------------------- #> 228.311 | 233.427 | 248.201 | 0.987 | 0.468 | 0.975 | 0.791 | 0.962- Instructions for interpeting each plot are in the subtitles
- Homogeneity of Variance shows a little bit of heterskedacity with the gentle slope from left to right.
- The Influential Observations doesn’t have any “contours,” but I assume the red points are the influential observations to be interested in.
- Normality of Residuals shows a bit of an issue in the right tail and maybe a little something in the left tail.
- The normality of REs by worker show all the point estimates within range with a few CIs outside.
- Overall this looks like a pretty good fit to me. Perhaps examine the influential observations. Also, the outcome variable shows some bi-modal-ness and left-skew, so maybe something could be done to model that better.
- Most of the metrics are for model comparison. ICC is over 0.10, R2(cond) is high so the modeling of the random effects is good. But R2 (marg) is quite low, so perhaps some transformations, interactions, splines, or just adding more adjustment variables can be raise that.
Example 2: {ggResidpanel}; Residuals vs Predictors
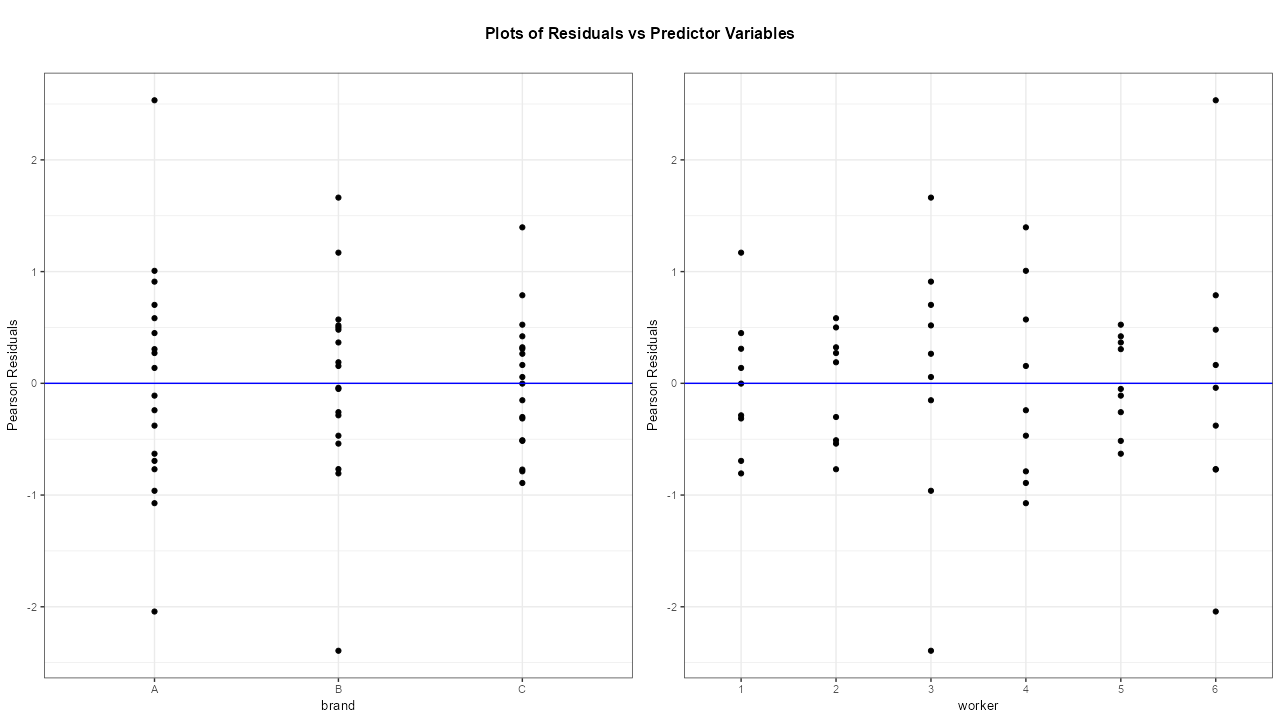
ggResidpanel::resid_xpanel(mod_bw_si)- Model from Example 1
- There are some large residuals in brands A and B and also for workers 3 and 6