General
Misc
- Resources
- Tools
- direnv - Augments existing shells with a new feature that can load and unload environment variables depending on the current directory
ctrl-rshell command history search- McFly - intelligent command history search engine that takes into account your working directory and the context of recently executed commands. McFly’s suggestions are prioritized in real time with a small neural network
- Path to a folder that’s above root folder:
- 1 level up:
../desired-folder - 2 levels up:
../../desired-folder
- 1 level up:
- Glob Patterns
- Notes from A Beginner’s Guide: Glob Patterns
- Patterns that can expand a wildcard pattern into a list of pathnames that match the given pattern.
- Wildcard Characters:
*,?, and[ ] - Negation of a Pattern:
!- e.g.
[!CBR]at
- e.g.
- Escape a character:
/ - Asterisk
*- On Linux, will match everything except slashes. On Windows, it will avoid matching backslashes as well as slashes.**- Recursively matches zero or more directories that fall under the current directory.*(pattern_list)- Only matches if zero or one occurrence of any pattern is included in the pattern-list above- Example: To find all Markdown files recursively that end with
.md, the pattern would be**/*.md*.mdwould only return the file paths in the current directory
- Question Mark
- Used to match any single character
- Case insensitive
- Useful for finding files with dates in the file name
- Example: To match files with the pattern “at” (e.g.
Cat.png), use?at- The
?is for the “C” or whatever the charater is preceding “at”
- The
- Square Brackets
- Character Classes
- Used to denote a pattern that should match a single character that is enclosed inside of the brackets. (“Character Classes” seems like a grand name for somethikng so simple.)
- The inside of the bracket can’t be empty (i.e. attempting to match a space)
- Case sensitive
- Example: To match
Cat.png,Bat.png, andRat.png, use[CBR]at
- Ranges
- Two characters that are separated by a dash.
- Example: Match all alpha numerical strings:
[A-Za-z0-9]
- Character Classes
R
Misc
The “shebang” line starting
#!allows a script to be run directly from the command line without explicitly passing it throughRscriptorr. It’s not required but is a helpful convenience on Unix-like systems.#!/usr/bin/env -S Rscript --vanilla- The shebang attempts to use
/usr/bin/envto locate theRscriptexecutable and then passes –vanilla as an argument toRscript
- The shebang attempts to use
Alter line endings when writing an R script on Windows but executing it on Linux
Windows uses \r\n (carriage return + newline) as line endings.
Linux/Unix uses \n (newline only) as line endings.
Command that makes the script compatible with Linux systems
sed -i 's/\r//' my-script.Rsed: Stream editor for filtering and transforming text.- -i: Edits the file “in place.”
- ‘s/\\r//’: Removes (s///) all occurrences of \r (carriage return).
- filename: The file to process.
Exiting R in bash without annoying “Save Workspace” prompt (source)
alias R="$(/usr/bin/which R) --no-save"
Resources
- Invoking R from the command line for using
RandR CMD
- Invoking R from the command line for using
Packages
- {ps} - List, Query, Manipulate System Processes
- {fs} - A cross-platform, uniform interface to file system operations
- Navarro deep dive, For fs
- {littler} - A scripting and command-line front-end for GNU R
- {rapp} - Package for building a polished CLI application from a simple R script
- An alternative front end to R, a drop-in replacement for Rscript that automatically parses command line arguments into R values
- {sys} - Offers drop-in replacements for the
system2with fine control and consistent behavior across platforms- Supports clean interruption, timeout, background tasks, and streaming STDIN / STDOUT / STDERR over binary or text connections.
- Arguments on Windows automatically get encoded and quoted to work on different locales.
- {seekr} - Recursively list files from a directory, filter them using a regular expression, read their contents, and extract lines that match a user-defined pattern
- Designed for quick code base exploration, log inspection, or any use case involving pattern-based file and line filtering.
File Paths
Various functions to make file paths easier to work with (source)
(path <- here::here()) #> [1] "C:/Users/ercbk/Documents" base::normalizePath(path) #> [1] "C:\\Users\\ercbk\\Documents" user <- "ercbk" base::file.path("C:Users", user, "Documents") #> [1] "C:Users/ercbk/Documents"here- A drop-in replacement forfile.paththat will always locate the files relative to your project root.normalizePath- Convert file paths to canonical form for the platformfile.path- (faster thanpaste) Construct the path to a file from components in a platform-independent way.
basename,dirname,tools::file_path_sans_extbasename("~/me/path/to/file.R") #> [1] "file.R" basename("C:\\Users\\me\\Documents\\R\\Projects\\file.R") #> [1] "file.R" dirname("~/me/path/to/file.R") #> [1] "~me/path/to" tools::file_path_sans_ext("~/me/path/to/file.R") #> [1] "~/me/path/to/file"
Rscriptneed to be onPATHRun R (default version) in the shell:
RS # or rig runRSmight require {rig} to be installed- To run a specific R version that’s already installed:
R-4.2
Run an R script:
Rscript "path\to\my-script.R" # or rig run -f <script-file> # or chmod +x my-script.R ./my-script.REvaluate an R expression:
Rscript -e <expression> # or rig run -e <expression>Run an R app:
rig run <path-to-app>- Plumber APIs
- Shiny apps
- Quarto documents (also with embedded Shiny apps)
- Rmd documents (also with embedded Shiny apps)
- Static web sites
Make an R script pipeable (From link)
parallel "echo 'zipping bin {}'; cat chunked/*_bin_{}_*.csv | ./upload_as_rds.R '$S3_DEST'/chr_'$DESIRED_CHR'_bin_{}.rds"#!/usr/bin/env Rscript library(readr) library(aws.s3) # Read first command line argument data_destination <- commandArgs(trailingOnly = TRUE)[1] data_cols <- list(SNP_Name = 'c', ...) s3saveRDS( read_csv( file("stdin"), col_names = names(data_cols), col_types = data_cols ), object = data_destination )- By passing
readr::read_csvthe function,file("stdin"), it loads the data piped to the R script into a dataframe, which then gets written as an .rds file directly to s3 using {aws.s3}.
- By passing
Killing a process
system("taskkill /im java.exe /f", intern=FALSE, ignore.stdout=FALSE)Kill all processes that are using a particular port (source)
pids <- system("lsof -i tcp:3000", intern = TRUE) |> readr::read_table() |> subset(select = "PID", drop = TRUE) |> unique() lapply(paste0("kill -9", pids), system)lsofis “List Open Files” by processes, users, and process IDs (source)-ilists open network connections and portstcp:3000specifies the type (TCP) and the port number (3000)- intern indicates whether to capture the output of the command as a character vector.
fuser -k 3000/tcpalso accomplishes this
Starting a process in the background
# start MLflow server sys::exec_background("mlflow server")Check file sizes in a directory
file.info(Sys.glob("*.csv"))["size"] #> size #> Data8277.csv 857672667 #> DimenLookupAge8277.csv 2720 #> DimenLookupArea8277.csv 65400 #> DimenLookupEthnic8277.csv 272 #> DimenLookupSex8277.csv 74 #> DimenLookupYear8277.csv 67- First one is about 800MB
Read first ten lines of a file
cat(paste(readLines("Data8277.csv", n=10), collapse="\n")) #> Year,Age,Ethnic,Sex,Area,count #> 2018,000,1,1,01,795 #> 2018,000,1,1,02,5067 #> 2018,000,1,1,03,2229 #> 2018,000,1,1,04,1356 #> 2018,000,1,1,05,180 #> 2018,000,1,1,06,738 #> 2018,000,1,1,07,630 #> 2018,000,1,1,08,1188 #> 2018,000,1,1,09,2157Delete an opened file in the same R session
You **MUST** unlink it before any kind of manipulation of object
- I think this works because readr loads files lazily by default
Example:
wisc_csv_filename <- "COVID-19_Historical_Data_by_County.csv" download_location <- file.path(Sys.getenv("USERPROFILE"), "Downloads") wisc_file_path <- file.path(download_location, wisc_csv_filename) wisc_tests_new <- readr::read_csv(wisc_file_path) # key part, must unlink before any kind of code interaction # supposedly need recursive = TRUE for Windows, but I didn't need it # Throws an error (hence safely) but still works safe_unlink <- purrr::safely(unlink) safe_unlink(wisc_tests_new) # manipulate obj wisc_tests_clean <- wisc_tests_new %>% janitor::clean_names() %>% select(date, geo, county = name, negative, positive) %>% filter(geo == "County") %>% mutate(date = lubridate::as_date(date)) %>% select(-geo) # clean-up fs::file_delete(wisc_file_path)
Find out which process is locking or using a file
- Open Resource Monitor, which can be found
- By searching for Resource Monitor or resmon.exe in the start menu, or
- As a button on the Performance tab in your Task Manager
- Go to the CPU tab
- Use the search field in the Associated Handles section
- type the name of file in the search field and it’ll search automatically
- 35548
- Open Resource Monitor, which can be found
List all file based on file names, directories, or glob patterns. (source)
list_all_files <- function(include = "*") { # list all files in the current directory recursing all_files <- fs::dir_ls(recurse = TRUE, all = TRUE, type = "any") # Identify directory patterns explicitly mentioned matched_dirs <- include[fs::dir_exists(include)] # Get *all* files inside matched directories extra_files <- unlist( lapply( matched_dirs, function(d) fs::dir_ls(d, recurse = TRUE, all = TRUE) ) ) # Convert glob patterns to regex regex_patterns <- sapply(include, function(p) { if (grepl("\\*", p)) { utils::glob2rx(p) # Convert glob to regex } else { paste0("^", p, "$") # Match exact filenames } }) # Match files against regex patterns matched_files <- all_files[sapply(all_files, function(file) { any(stringr::str_detect(file, regex_patterns)) })] # Combine matched files and all directory contents unique(c(matched_files, extra_files)) } include <- c("*.Rmd", "*.tex", "_book", "_output.yml", "\\.R$", "thumbnail.jpg", 'README.*') list_all_files(include = include)
Python
Notes from Python’s many command-line utilities
- Lists and describes all CLI utilities that are available through Python’s standard library
Packages
Linux utilities through Python in CLI
Command Purpose More python3.12 -m uuidLike uuidgenCLI utilityDocs python3.12 -m sqlite3Like sqlite3CLI utilityDocs python -m zipfileLike zip&unzipCLI utilitiesDocs python -m gzipLike gzip&gunzipCLI utilitiesDocs python -m tarfileLike the tarCLI utilityDocs python -m base64Like the base64CLI utilitypython -m ftplibLike the ftputilitypython -m smtplibLike the sendmailutilitypython -m poplibLike using curlto read emailpython -m imaplibLike using curlto read emailpython -m telnetlibLike the telnetutilityuuidandsqlite3require version 3.12 or above.
Code Utilities
Command Purpose More python -m pipInstall third-party Python packages Docs python -m venvCreate a virtual environment Docs python -m pdbRun the Python Debugger Docs python -m unittestRun unittesttests in a directoryDocs python -m pydocShow documentation for given string Docs python -m doctestRun doctests for a given Python file Docs python -m ensurepipInstall pipif it’s not installedDocs python -m idlelibLaunch Python’s IDLE graphical REPL Docs python -m zipappTurn Python module into runnable ZIP Docs python -m compileallPre-compile Python files to bytecode Docs pathlib
/=Operator for joining pathsfrom pathlib import Path directory = Path.home() directory /= "Documents" directory #> PosixPath('/home/trey/Documents')
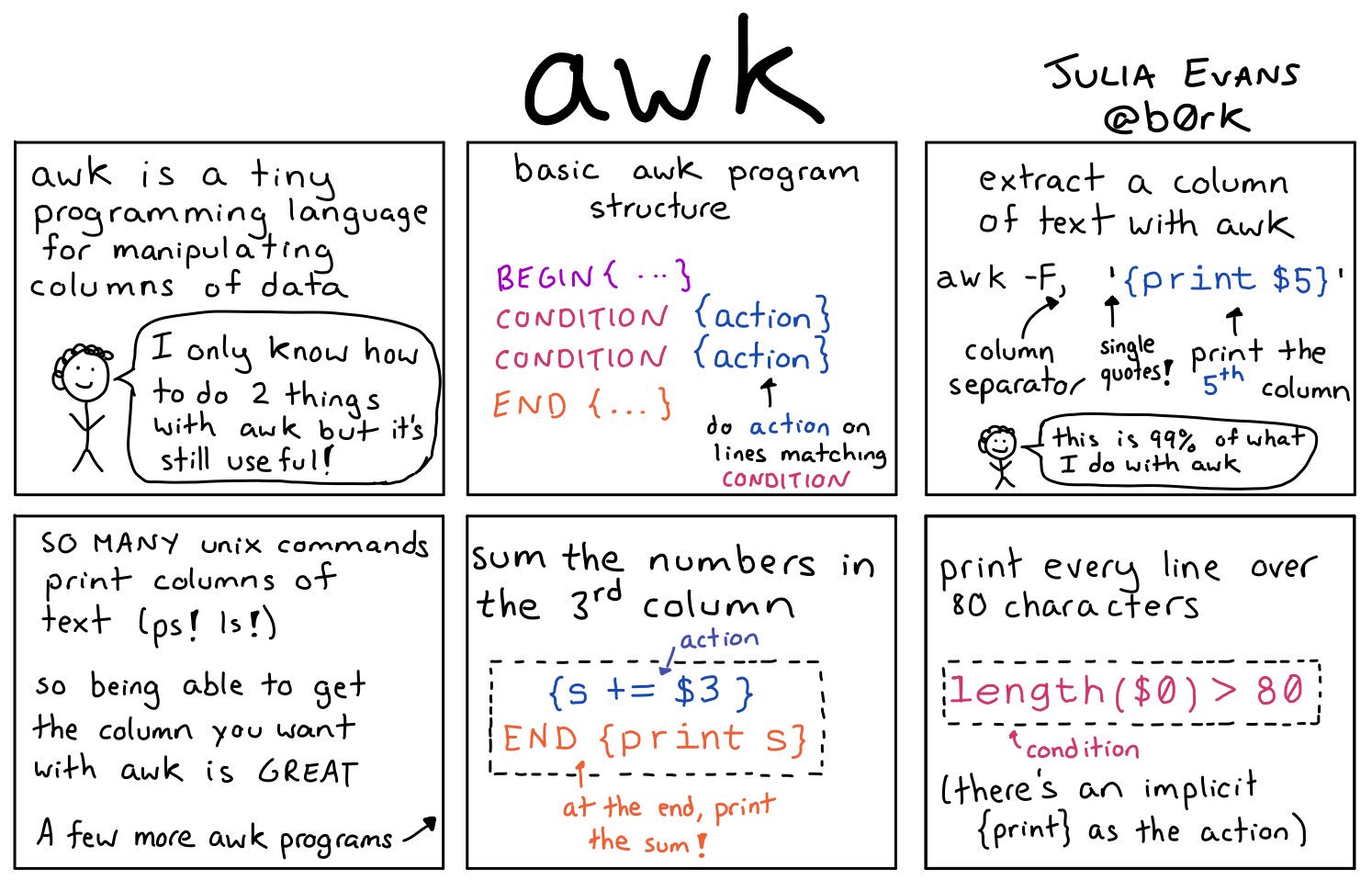
AWK
Misc
- Resources
Print first few rows of columns 1 and 2
awk -F, '{print $1,$2}' adult_t.csv|headExtract every 4th line starting from the line 1 (i.e. 1, 5, 9, 13, …)
awk '(NR%4==1)' file.txtFilter lines where no of hours/ week (13th column) > 98
awk -F, ‘$13 > 98’ adult_t.csv|headFilter lines with “Doctorate” and print first 3 columns
awk '/Doctorate/{print $1, $2, $3}' adult_t.csvRandom sample 8% of the total lines from a .csv (keeps header)
'BEGIN {srand()} !/^$/ {if(rand()<=0.08||FNR==1) print > "rand.samp.csv"}' big_fn.csvDecompresses, chunks, sorts, and writes back to S3 (From link)
# Let S3 use as many threads as it wants aws configure set default.s3.max_concurrent_requests 50 for chunk_file in $(aws s3 ls $DATA_LOC | awk '{print $4}' | grep 'chr'$DESIRED_CHR'.csv') ; do aws s3 cp s3://$batch_loc$chunk_file - | pigz -dc | parallel --block 100M --pipe \ "awk -F '\t' '{print \$1\",...\"$30\">\"chunked/{#}_chr\"\$15\".csv\"}'" # Combine all the parallel process chunks to single files ls chunked/ | cut -d '_' -f 2 | sort -u | parallel 'cat chunked/*_{} | sort -k5 -n -S 80% -t, | aws s3 cp - '$s3_dest'/batch_'$batch_num'_{}' # Clean up intermediate data rm chunked/* done
Vim
- Command-line based text editor
- Common Usage
- Edit text files while in CLI
- Logging into a remote machine and need to make a code change there. vim is a standard program and therefore usually available on any machine you work on.
- When running
git commit, by default git opens vim for writing a commit message. So at the very least you’ll want to know how to write, save, and close a file.
- Resources
- 2 modes: Navigation Mode; Edit Mode
- When Vim is launched you’re in Navigation mode
- Press i to start edit mode, in which you can make changes to the file.
- Press Esc key to leave edit mode and go back to navigation mode.
- Commands
xdeletes a characterdddeletes an entire rowb(back) goes to the previous wordn(next) goes to the next word:wqsaves your changes and closes the file:q!ignores your changes and closes the filehis \(\leftarrow\)jis \(\downarrow\)kis \(\uparrow\)l(i.e. lower L) is \(\rightarrow\)