GLMM
Misc
- Also see Mixed Effects, General >> Misc >> Packages
- {lme4}, {multilevelmod} which have some glm support
- {mixedup} for extracting components
- {glmmTMB}
- Beta, Beta-Binomial, Beta-Ordinal, various extensions for Negative Binomial and Poisson, Log-Normal, Tweedie, Student-T, Gamma, Zero-Inflated, Truncated (See Manual >> nbinom2 for details)
- Fixed and random effects models can be specified for the conditional and zero-inflated components of the model, as well as fixed effects for the dispersion parameter.
- Currently has an experimental implementation of fixed effect priors which can be a solution to complete separation errors, singular fits, and instability (Link).
- Supports parallelization through OpenMP but might not be supported by Mac and has significant overhead on Windows.
- {GLMMadaptive} - For binary/dichotomous data and count data with small counts and few repeated measurements, the accuracy of Laplace approximations (i.e. {glmmTMB}) is low. This package uses adaptive Gaussian quadrature rule which is the recommended, albeit more computationally intensive, approximation method for MLE in this situation.
- Supported Distribution Families: Student’s-t, Beta, Zero-Inflated and Hurdle Poisson and Negative Binomial, Hurdle Log-Normal, Hurdle Beta, Gamma, and Censored Normal
- Currently no nested or crossed random effects designs
- Can penalize fixed effects with a student-t prior
- Parallelization through {optimParallel} which doesn’t seem to have any OS limitations.
- {glmmPen} (Vignette)
- Fits high dimensional penalized generalized linear mixed models using the Monte Carlo Expectation Conditional Minimization (MCECM) algorithm.
- Performs variable selection on both the fixed and random effects simultaneously through MCP, SCAD, or LASSO penalties.
- Supports Binomial, Gaussian, and Poisson data with canonical links
- {mgcv::gamm} - Fits the specified generalized additive mixed model (GAMM) to data, by a call to {nlme::lme} in the normal errors identity link case, or by a call to
gammPQL(a modification of glmmPQL from {MASS}) otherwise.- ** There is a bug (as of Jan 2025) in
gammthat prohibits the random effects from being included in the fitted values when the random argument is used and there’s not a spline in the formula ** (See Examples >> Example 1)- In docs for the function, it does say that a manual calculation is required, but I’ve only found that to be the case in the specification stated above.
- Performs poorly with binary data, since it uses PQL. It is better to use
gamwiths(...,bs="re")terms, or {gamm4}. - Usually much slower than
gam - Correlation Structures: The routine will be very slow and memory intensive if correlation structures are used for the very large groups of data.
- e.g. Attempting to run the spatial example in the examples section with many 1000’s of data is definitely not recommended: often the correlations should only apply within clusters that can be defined by a grouping factor, and provided these clusters do not get too huge then fitting is usually possible.
- ** There is a bug (as of Jan 2025) in
- {gamm4} - Generalized Additive Mixed Models using {mgcv} and {lme4}
- Based on
gammfrom mgcv, but uses lme4 rather than nlme as the underlying fitting engine - More robust numerically than
gamm, and by avoiding PQL gives better performance for binary and low mean count data. - Most useful when:
- The random effects are not i.i.d.
- There are large numbers of random coeffecients (more than several hundred), each applying to only a small proportion of the response data.
- Tensor product smoothing is available via t2 terms
- Issues
- It can not handle most multi-penalty smooths (i.e. not te type tensor products or adaptive smooths)
- No facilty for nlme style correlation structures.
- For fitting GAMMs with modest numbers of i.i.d. random coefficients then gamm4 is slower than gam (or bam for large data sets)
- Based on
- {glmertree} - Generalized Linear Mixed Model Trees
- Combines
lmer/glmerfrom {lme4} andlmtree/glmtreefrom {partykit}
- Combines
- Papers
- High performance implementation of the hierarchical likelihood for generalized linear mixed models: an application to estimate the potassium reference range in massive (codelectronic health records datasets
- A high-performance, direct implementation of the hierarchical-likelihood for GLMMs in the R package TMB
- The hierarchical likelihood approach to GLMMs is a methodologically rigorous framework for the estimation of GLMMs that is based on the Laplace Approximation (LA), which replaces integration with numerical optimization, and thus scales very well with dimensionality.
- Code (+ parallelization option)
SingleRE: Analyzes a Poisson GLM with a single random effectMultipleRE: Analyzes a Poisson GLM with multiple random effects
- High performance implementation of the hierarchical likelihood for generalized linear mixed models: an application to estimate the potassium reference range in massive (codelectronic health records datasets
Model Equation
- Equation
\[ g\{E(y_i \mid b_i)\} = \beta X_i + b_i Z_i, \]
- \(g(⋅)\): A monotonic link function
- \(X_i\): A design matrix for the fixed effects
- \(\beta\): Fixed effects coefficients
- \(Z_i\): A design matrix for the random effects that’s assumed to be a subset of \(X_i\)
- \(b_i\): Random effects coefficients
Examples
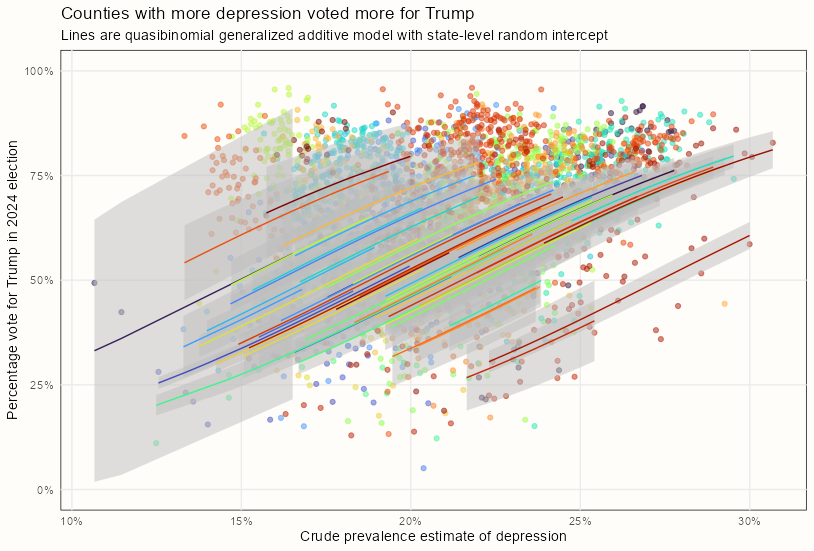
Example 1: GAMM with Varying Intercepts (source, source)
- Did counties with more depressed people voted more for Trump?
- Binomial and Quasi-Binomial distributions are used to model a percent response variable
library(tidyverse) # https://stacks.cdc.gov/view/cdc/129404 dep <- readxl::read_excel("cdc_129404_DS1.xlsx", skip = 1) election_data_url <- "https://raw.githubusercontent.com/tonmcg/US_County_Level_Election_Results_08-24/refs/heads/master/2024_US_County_Level_Presidential_Results.csv" votes <- read_csv(election_data_url) dat_votes_dep <- votes |> inner_join(dep, by = c("county_fips" = "CountyFIPS code")) |> mutate(cpe = `Crude Prevalence Estimate` / 100, aape = `Age-adjusted Prevalence Estimate` / 100) glimpse(votes_dep_dat) #> Rows: 3,107 #> Columns: 18 #> $ state_name <chr> "Alabama", "Alabama", "Alabama", "Alabama", "Alabama", "Alabama", "Alaba… #> $ county_fips <chr> "01001", "01003", "01005", "01007", "01009", "01011", "01013", "01015", … #> $ county_name <chr> "Autauga County", "Baldwin County", "Barbour County", "Bibb County", "Bl… #> $ votes_gop <dbl> 20447, 95144, 5578, 7563, 25271, 1099, 5167, 34841, 8704, 11342, 16901, … #> $ votes_dem <dbl> 7429, 24763, 4120, 1617, 2569, 2983, 3248, 13170, 5402, 1550, 2697, 2511… #> $ total_votes <dbl> 28139, 120973, 9766, 9230, 28024, 4104, 8459, 48435, 14215, 12965, 19708… #> $ diff <dbl> 13018, 70381, 1458, 5946, 22702, -1884, 1919, 21671, 3302, 9792, 14204, … #> $ per_gop <dbl> 0.7266427, 0.7864895, 0.5711653, 0.8193933, 0.9017628, 0.2677875, 0.6108… #> $ per_dem <dbl> 0.26401080, 0.20469857, 0.42187180, 0.17518960, 0.09167142, 0.72685185, … #> $ per_point_diff <dbl> 0.46263193, 0.58179098, 0.14929347, 0.64420368, 0.81009135, -0.45906433,… #> $ CountyName <chr> "Autauga", "Baldwin", "Barbour", "Bibb", "Blount", "Bullock", "Butler", … #> $ `State Name` <chr> "Alabama", "Alabama", "Alabama", "Alabama", "Alabama", "Alabama", "Alaba… #> $ `Crude Prevalence Estimate` <dbl> 24.52297, 24.61480, 23.68052, 24.80906, 26.65236, 22.00814, 23.88319, 26… #> $ `95% Confidence Interval for Crude Estimates` <chr> "23.3-25.8", "23.3-26", "22.7-24.8", "23.6-26.1", "25.3-28.1", "21-23.1"… #> $ `Age-adjusted Prevalence Estimate` <dbl> 24.9, 25.7, 24.2, 25.1, 27.5, 22.4, 24.9, 27.0, 24.9, 27.5, 27.6, 25.3, … #> $ `95% Confidence Interval for Age-adjusted Estimates` <chr> "23.6-26.4", "24.3-27.1", "23.1-25.3", "23.9-26.4", "26-29", "21.3-23.6"… #> $ cpe <dbl> 0.2452297, 0.2461480, 0.2368052, 0.2480906, 0.2665236, 0.2200814, 0.2388… #> $ aape <dbl> 0.249, 0.257, 0.242, 0.251, 0.275, 0.224, 0.249, 0.270, 0.249, 0.275, 0.…- per_gop: Percentage of votes cast for the Republican presidential candidate
- cpe: Crude Prevalence Estimate (percentage); depression measure
mod_bin_inter <- lme4::glmer(per_gop ~ cpe + (1 | state_name), family = "binomial", weights = total_votes, data = dat_votes_dep) summary(mod_bin_inter) #> Random effects: #> Groups Name Variance Std.Dev. #> state_name (Intercept) 0.4636 0.6809 #> Number of obs: 3107, groups: state_name, 50 #> #> Fixed effects: #> Estimate Std. Error z value Pr(>|z|) #> (Intercept) -3.27338 0.09572 -34.2 <2e-16 *** #> cpe 16.40275 0.01073 1529.1 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Correlation of Fixed Effects: #> (Intr) #> cpe -0.022- This lets the vote for Trump in any state vary for all the state-specific things that aren’t in our model.
- States weighted based on their overall number of voters, which makes particular sense for a binomial or similar family response.
- {lme4} doesn’t support a quasi-binomial distribution. With the binomial distribution, it’s assumed that the relationship between the mean and variance is \(p(1-p)/n\) which means overdispersion isn’t being handled correctly.
- This results in CIs that are too narrow.
- We see an overall (constant nation-wide) relationship (fixed effect) between depression (cpe) and voting (per_gop) in the counties (observation-level) within each state.
# must be a factor to use as a random effect in gam dat_votes_dep <- mutate(dat_votes_dep, state_name = as.factor(state_name)) # These two specifications included random effects in fitted values mod_quasi_bin_inter_gam <- mgcv::gam(per_gop ~ cpe + s(state_name, bs = 're'), weights = total_votes, family = quasibinomial, data = dat_votes_dep) mod_quasi_bin_inter_spl <- mgcv::gamm(per_gop ~ cpe + s(state_name, bs = 're'), weights = total_votes, family = quasibinomial, data = dat_votes_dep) # This specification does NOT include randome effects in fitted values # mod_quasi_bin_inter_lme <- # mgcv::gamm(per_gop ~ cpe, # random = list(state_name = ~ 1), # weights = total_votes, # family = quasibinomial, # data = dat_votes_dep) summary(mod_quasi_bin_inter_spl$gam) #> Parametric coefficients: #> Estimate Std. Error t value Pr(>|t|) #> (Intercept) -3.1087 0.1392 -22.33 <2e-16 *** #> cpe 15.7091 0.5869 26.77 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Approximate significance of smooth terms: #> edf Ref.df F p-value #> s(state_name) 45.65 49 20.99 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> R-sq.(adj) = 0.346 #> Scale est. = 3193.2 n = 3107 broom::glance(mod_quasi_bin_inter_gam) #> # A tibble: 1 × 9 #> df logLik AIC BIC deviance df.residual nobs adj.r.squared npar #> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <int> #> 1 50.4 NA NA NA 10024845. 3057. 3107 0.345 52{mgcv} does have a quasi-binomial family available
Either the
gamfunction or thegammfunction can be used to fit this model by usings(state_name, bs = "re"). It should NOT be fit usinggammand the random argument, unless you aren’t interested the fitted values or you want to include the random effects manually.- Using the California data as my test case, I also noticed that using
gammwithout a spline produced negative fitted values which I don’t think should be possible with a Quasi-Binomial.
- Using the California data as my test case, I also noticed that using
gammvs.gam(with spline “re” specification)- Even though there are differences, these are practically the same model
- There are fewer effective degrees of freedom (edf) used to fit the spline (i.e. smoother curve) using
gammthan withgam(45.65 vs 48.44). - The effect size is slightly lower using
gammvsgam(15.7091 vs 16.3333) - The standard error is slightly lower using
gammvsgam(0.5869 vs 0.6224) - According to the docs,
gamshould be the faster fitting function
The standard error for cpe is much higher 0.5869, compared to 0.0107 in the Binomial model
- This will result in fatter, more realistic CI intervals
-
Code
# using gam aug_quasi_bin_inter_gam <- broom::augment(mod_quasi_bin_inter, type.predict = "response") |> mutate(lower = .fitted - 1.96 * .se.fit, upper = .fitted + 1.96 * .se.fit) # using gamm pred_quasi_bin_inter_spl <- predict(mod_quasi_bin_inter_spl$gam, se.fit = TRUE, type = "response") aug_quasi_bin_inter_spl <- dat_votes_dep |> mutate(.fitted = pred_quasi_bin_inter_spl$fit, lower = .fitted - 1.96 * pred_quasi_bin_inter_spl$se.fit, upper = .fitted + 1.96 * pred_quasi_bin_inter_spl$se.fit) |> select(per_gop, cpe, state_name, .fitted, lower, upper) ggplot(aug_quasi_bin_inter_gam, aes(x = cpe, group = state_name)) + geom_point(aes(y = per_gop, color = state_name), alpha = 0.5) + geom_ribbon(aes(ymin = lower, ymax = upper), fill = "gray", alpha = 0.5) + geom_line(aes(y = .fitted, color = state_name)) + scale_y_continuous(limits = c(0, 1), label = scales::label_percent()) + scale_x_continuous(label = scales::label_percent()) + scale_color_viridis_d(option = "turbo") + labs(x = "Crude prevalence estimate of depression", y = "Percentage vote for Trump in 2024 election", subtitle = "Lines are quasibinomial generalized additive model with state-level random intercept", title = "Counties with more depression voted more for Trump") + theme_notebook(legend.position = "none")
mod_quasi_bin_inter_spat_lme <- mgcv::gamm(per_gop ~ cpe + s(long, lat), random = list(state_name = ~1), weights = total_votes, family = quasibinomial, data = dat_votes_dep_spat) summary(mod_quasi_bin_inter_spat_lme) #> Parametric coefficients: #> Estimate Std. Error t value Pr(>|t|) #> (Intercept) -2.8878 0.1481 -19.50 <2e-16 *** #> cpe 15.0166 0.6271 23.95 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Approximate significance of smooth terms: #> edf Ref.df F p-value #> s(long,lat) 25.41 25.41 7.129 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> R-sq.(adj) = 0.0981 #> Scale est. = 3005.3 n = 3107 AIC(mod_quasi_bin_inter_spat_lme$lme, mod_quasi_bin_inter_spl$lme) #> df AIC #> mod_quasi_bin_inter_spat_lme$lme 7 8806.902 #> mod_quasi_bin_inter_spl$lme 4 8879.840With a spline function in the formula, there will now be random effects included with the fitted values when using
gammwith random argument- I assume this is a bug.
The hope was to account for some of the probable spatial autorcorrelation. The R2 indicates an even worse fit while potentially more predictive given the lower AIC.
- See Mixed Effects, Residual Covariance Structures >> Examples >> Spatial Autocorrelation >> Example 1
Note that the data used in this model is dat_votes_dep_spat which includes the lat and long predictors that represent centroids for each county. Code is included below.
-
Code
pred_quasi_bin_inter_spat_lme <- predict(mod_quasi_bin_inter_spat_lme$gam, se.fit = TRUE, type = "response") aug_quasi_bin_inter_spat_lme <- dat_votes_dep_spat |> mutate(.fitted = pred_quasi_bin_inter_spat_lme$fit, lower = .fitted - 1.96 * pred_quasi_bin_inter_spat_lme$se.fit, upper = .fitted + 1.96 * pred_quasi_bin_inter_spat_lme$se.fit) |> select(per_gop, cpe, state_name, .fitted, lower, upper) ggplot(aug_quasi_bin_inter_spat_lme, aes(x = cpe, group = state_name)) + geom_point(aes(y = per_gop, color = state_name), alpha = 0.5) + geom_ribbon(aes(ymin = lower, ymax = upper), fill = "black", alpha = 0.5) + scale_y_continuous(limits = c(0, 1), label = scales::label_percent()) + scale_x_continuous(label = scales::label_percent()) + scale_color_viridis_d(option = "turbo") + facet_wrap(~state_name) + labs(x = "Crude prevalence estimate of depression", y = "Percentage vote for Trump in 2024 election", subtitle = "Gray ribbons are 95% confidence intervals from quasibinomial generalized additive model with spatial effect and state-level random intercept effect", title = "Counties with more depression voted more for Trump") + theme_notebook(legend.position = "none")- A closer look at the black ribbons in these plots will show some states have some really squiggly lines (overfitting). This must be due to the longitude and latitude spline. I attempted to reduce the number on knots, and even at k = 2, some ribbons remained very squiggly. I’m not sure why some of these lines werent’ affected. I didn’t drill down into the individual states data and fitted values further to see what was going on.
- Some state’s data and their trend lines (ribbons) didn’t seem to match either.
Centroid Calculations
Code
library(sf) download.file('https://www2.census.gov/geo/tiger/GENZ2023/shp/cb_2023_us_county_500k.zip', destfile = "../../Data/cb_2023_us_county_500k.zip", mode = "wb") unzip("../../Data/cb_2023_us_county_500k.zip", exdir = "../../Data") counties <- st_read("../../Data/cb_2023_us_county_500k.shp") county_cent <- counties |> st_centroid() sc <- st_coordinates(county_cent) county_cent <- county_cent |> mutate(long = sc[, 1], lat = sc[, 2], # combine the two digit state code with the 3 digit county code: county_fips = paste0(STATEFP, COUNTYFP)) dat_votes_dep_spat <- dat_votes_dep |> left_join(county_cent, by = "county_fips")
Example 2: GAMM with Varying Intercepts and Varying Slopes (source)
mod_quasi_bin_intslp_spat <- mgcv::gamm(per_gop ~ cpe + s(long, lat), random = list(state_name = ~1 + cpe), weights = total_votes, family = quasibinomial, data = dat_votes_dep_spat summary(mod_quasi_bin_intslp_spat$gam) #> Parametric coefficients: #> Estimate Std. Error t value Pr(>|t|) #> (Intercept) -2.0995 0.5203 -4.035 5.59e-05 *** #> cpe 10.4828 2.5033 4.188 2.90e-05 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Approximate significance of smooth terms: #> edf Ref.df F p-value #> s(long,lat) 26.15 26.15 8.248 <2e-16 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> R-sq.(adj) = 0.186 #> Scale est. = 2597.3 n = 3107 AIC(mod_quasi_bin_intslp_spat$lme, mod_quasi_bin_inter_spat_lme$lme) #> df AIC #> mod_quasi_bin_intslp_spat$lme 9 8605.691 #> mod_quasi_bin_inter_spat_lme$lme 7 8806.902- This is a continuation from Example 1 where mod_quasi_bin_inter_spat_lme is a varying intercepts only model
- This is an overall better model — better fit (higher R2) and more predictive (lower AIC)
- Development of this model continues at Mixed Effects, Residual Covariance Structures >> Examples >> Spatial Autocorrelation >> Example 1
- Visualization code can be found in Example 1