Time Series
Misc
- Also see EDA, Time Series >> Association
- Packages
- {tseriesEntropy} - Implements an Entropy measure of dependence based on the Bhattacharya-Hellinger-Matusita distance.
Srho.test- Entropy Test For Serial And Cross Dependence For Categorical SequencesSrho.ts- Entropy based measure of serial and cross dependence for continuous data. Implements a normalized version of the Hellinger/Matusita distance
- {SimIndep} - Weighted Similarity Aggregation Test for Serial Independence
- Particularly suited for high-dimensional and non-Euclidean time series
- {stochcorr} - Stochastic Correlation Modelling via Circular Diffusion
- Useful for measuring time series coorelation where there’s random noise associated with both series (e.g. stock prices)
- {wqc} - Estimate and plot wavelet quantile correlations between two time series. Wavelet quantile correlation is used to capture the dependency between two time series across quantiles and different frequencies.
- {tseriesEntropy} - Implements an Entropy measure of dependence based on the Bhattacharya-Hellinger-Matusita distance.
- Papers
- Measuring General Associations in Time Series: An Adaptation and Empirical Evaluation of the CODEC Coefficient in Determining Autoregressive Dynamics
- Utilizes the FOCI feature selection algorithm (extension of Chatterjee’s \(\xi\)) to select lags (See Feature Selection >> More Advanced >> FOCI).
- Demonstrated improved detection of significant lags even without pre-processing steps such as differencing or additive decomposition especially for time series that are non-stationary or exhibits nonlinear relationships.
- Has weak performance in time series characterized by conditional heteroskedasticity, as observed in GARCH-type models
- The first lag identified was sensitive to outliers and extreme values. The second and third lags identified demonstrated greater robustness in estimating p, although they exhibited bias in most of the scenarios considered.
- Measuring General Associations in Time Series: An Adaptation and Empirical Evaluation of the CODEC Coefficient in Determining Autoregressive Dynamics
- Schwert’s Rule for the maximum number of lags to be considered (Paper)
\[ h^* = 12 \left(\frac{n}{100}\right)^{1/4} \] - Granger Causality - Determines whether one time series is useful (i.e. containing information beyond only using the lagged series of the target series) in forecasting another. Assumes stationarity and linear association.
Cross-Correlation Function (CCF)
The correlation between two stationary series. The cross-correlation function (CCF) helps you determine which lags of time series \(X\) predicts the value of time series \(Y\).
For a set of sample correlations between \(x_{t+h}\) and \(y_t\) for \(h = 0, \pm1, \pm2, \pm3, \ldots\), a negative value for \(h\) is a correlation between \(x\) at a time before \(t\) (i.e. lag) and \(y\) at time \(t\).
- For instance, consider \(h = −2\). The CCF value would give the correlation between \(x_{t−2}\) and \(y_t\).
Misc
- Notes from
- Time Series Analysis With Applications in R by Cryer and Chan; (pp. 260-271)
- Packages
- {ebtools::prewhitened_ccf}
- {forecast::ggCcf} - You have to manually prewhiten series before using this function.
- {feasts::CCF} - You have to manually prewhiten series before using this function. Data needs to be a tsibble.
- Resources
- Time Series Analysis and Its Applications by Shumway and Stoffer (pp. 18, 23)
- When calculating correlations between lags of a variable and the variable itself (ACF) or another variable (CCF) you can NOT simply take corr(xt-k, yt) for the correlation of x at lag k and y and repeat for all the lags of x.
- The correlation formula/function requires the mean of the series, you would be using a different mean for xk for each calculation of the correlation of each pair. The assumption is that the series are (second-order?) stationary and therefore have a constant mean (i.e. each series has 1 mean (and variance)), so the mean(s) of (each) original series should be used in the calculations of the correlations.
- Source: Modern Applied Statistics with S, Venables and Ripley
- Compared one against the other in COVID-19 CFR project and they actually produce similar patterns but different values of the CCF. The simple correlation method produced inflated CCF values.
- Notes from
Terms
- Input and Output Series - In a cross-correlation in which the direction of influence between two time-series is hypothesized or known,
- The influential time-series is called the “input” time-series
- The affected time-series is called the “output” time-series.
- Lead - When one or more \(x_{t+h}\) , with \(h\) negative, are predictors of \(y_t\), it is sometimes said that x leads y.
- Lag - When one or more \(x_{t+h}\), with \(h\) positive, are predictors of \(y_t\), it is sometimes said that x lags y.
- Input and Output Series - In a cross-correlation in which the direction of influence between two time-series is hypothesized or known,
Prewhitening
- If either series contain autocorrelation, or the two series share common trends, it is difficult for the CCF to identify meaningful relationships between the two time series. Pre-whitening solves this problem by removing the autocorrelation and trends.
- Helps to avoid spurious correlations
- Make sure there’s a theoretical reason for the two series to be related
- Autocorrelation of at least one series should be removed (transformed to white noise)
- With autocorrelation present:
- The variance of cross-correlation coefficient is high and therefore spurious correlations are likely
- Significance calculations are no longer valid since the CCF distribution will not be normal and the variance is no longer 1/n
- With autocorrelation present:
- A problem even with stationary series (even more so with non-stationary series)
- We expect about 1.75 false alarms out of the 35 sample cross-correlations even after prewhitening
Steps
- Test for stationarity
- Find number of differences to make the series stationary
- Not sure if each series should be differenced the same number of times
- Why would you test a seasonal series and non-seasonal series for an association?
- The requirement is stationarity, so maybe try using the highest difference needed between both series
- In dynamic regression, Hyndman says difference all variables if one needs differencing, so proabably applicable here.
- Some examples also used log transformation , but when I did, it produced nonsensical CCF values. (covid cfr project). So, beware.
- Not sure if each series should be differenced the same number of times
- Seasonal Difference before differencing for trend
- (Optional) Create lag scatter plots of the differenced series and look for patterns to get an idea of the strength of the linear correlation (See EDA, Time Series >> Association)
- If there’s a nonlinear pattern, might be difficult or might not to use a nonlinear or nonparametric correlation function. See 3rd bullet under CCF header above for discussion of stationarity assumption. May not be necessary with another correlation type.
- Prewhiten both series (cryer, chan method)
- Apply correlation function
Example COVID-19 CFR project
# Number of differences ind_cases_diff <- forecast::ndiffs(ind_cases_ts) ind_deaths_diff <- forecast::ndiffs(ind_deaths_ts) # Number of seasonal differences ind_cases_sdiff <- forecast::nsdiffs(ind_cases_ts) ind_deaths_sdiff <- forecast::nsdiffs(ind_deaths_ts) # Only detrending differences; No seasonal diffs needed ind_cases_proc <- diff(ind_cases_ts, ind_cases_diff) ind_deaths_proc <- diff(ind_deaths_ts, ind_deaths_diff) # Fit AR model with processed input series ind_cases_ar <- ind_cases_proc %>% ts_tsibble() %>% model(AR(value ~ order(p = 1:30), ic = "aicc")) # Pull AR coefs ind_ar <- coef(ind_cases_ar) %>% filter(stringr::str_detect(term, "ar")) %>% pull(estimate) # Linearly filter both input and output series using coefs ind_cases_fil <- stats::filter(ind_cases_proc, filter = c(1, -ind_ar), method = 'convolution', sides = 1) ind_deaths_fil <- stats::filter(ind_deaths_proc, filter = c(1, -ind_ar), method = 'convolution', sides = 1) # spike at -20 with corr = 0.26; nonsensical lags at -4 and -15, -68 ggCcf(ind_cases_fil, ind_deaths_fil)
Distances
- The smaller the total distance between two series, the greater the association.
- Distance algorithms in the {dtw} and {dtwclust} packages
- Other packages
- {distantia} - Qantifies dissimilarity between time series
- Types of ts
- Multivariate and univariate time series.
- Regular and irregular sampling.
- Time series of different lengths.
- Features
- 10 distance metrics
- The normalized dissimilarity metric psi.
- Free and Restricted Dynamic Time Warping (DTW) for shape-based comparison.
- A Lock-Step method for sample-to-sample comparison
- Restricted permutation tests for robust inferential support.
- Analysis of contribution to dissimilarity of individual variables in multivariate time series.
- Hierarchical and K-means clustering of time series based on dissimilarity matrices.
- Types of ts
- {IncDTW} - Fast DTW in C++; Incremental Calculation of Dynamic Time Warping
- Incremental calculation of DTW (reduces runtime complexity to a linear level for updating the DTW distance)
- Vector-based implementation of DTW which is faster because no matrices are allocated (reduces the space complexity from a quadratic to a linear level in the number of observations)
- Subsequence matching algorithm runDTW efficiently finds the k-NN to a query pattern in a long time series
- {twdtw} - Implements Time-Weighted Dynamic Time Warping (TWDTW), a measure for quantifying time series similarity.
- Applicable to multi-dimensional time series of various resolutions. It is particularly suitable for comparing time series with seasonality for environmental and ecological data analysis
- {distantia} - Qantifies dissimilarity between time series
- Also see Clustering, Time Series >> {dtwclust}
Dynamic Time Warping Distance (DTW)
dtw_basic,dtw2- The distance between 2 series is measured by “warping” the time axis to bring them as close as possible to each other and measuring the sum of the distances between the points.
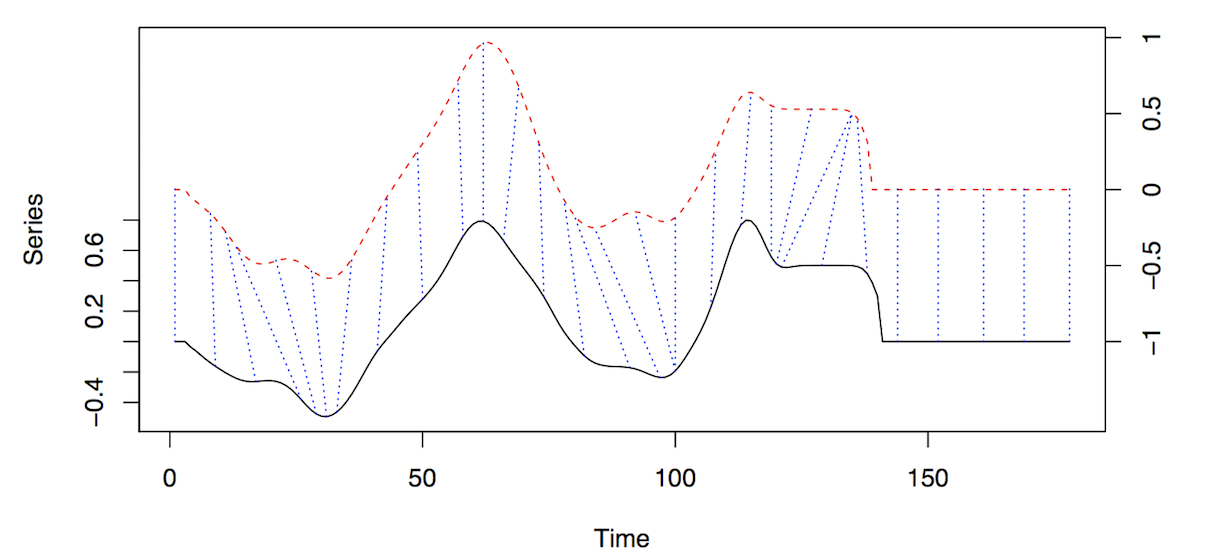
- Figure shows alignment of two series, x and y.
- The initial and final points of the series must match, but other points along the axis may be “warped” in order to minimize the “cost” (i.e. weighted distance).
- The dashed blue lines are the warp curves and show how points are mapped to each other.
- Figure shows alignment of two series, x and y.
- Hyperparameters:
- Step Pattern - Determines how the algorithm traverses the rows of the LCM to find the optimal path (See Step Patterns Section)
- Window Range - Limits the number lcm calculations for each point (See Window Constraint Section)
- Steps
- Calculate LCM matrix for series X and Y where X is the Query or Test Series and Y is the Reference Series. The LCM is a distance matrix where each element is the distance between the ith point of series 1 and the jth point of series 2. (See Local Cost Matrix (LCM) for details)
- Simultaneously move along each row of the LCM using a chosen step pattern within a chosen window (See Step Patterns and Window Constraints sections to get a visual of this process).
The minimum lcm for each point along x-axis is found. The sequence of minimum lcms or minimum alignment is \(\phi\) .
Calculate the cost, DTWp, using the LCMs in the minimum alignment
\[ \text{DTW}_p(x, y) = \left(\sum\frac{m_\phi \text{lcm}(k)^p}{M_\phi}\right)^{1/p} \]
- \(m_\phi\): Per-step weighting coefficient (edge weight in patterns fig)
- \(M_\phi\): Normalization constant
- \(k\): Pair of points (or position along the x-axis) in the minimum alignment
dtw_basicsets \(p = 2\)- The
dtwfunction in the dtw package doesn’t use p in this equation
- The
- Choose the alignment with the lowest cost, \(\text{DTW}_p\) (i.e. sum of lcm distances for that alignment)
Local Cost Matrix (LCM)
Computed for each pair of series that are compared
The \(L_p\) norm (i.e. distance) between the query series and reference series
\[ \text{lcm}(i, j) = \left(\sum_v | x_i^v - y_j^v|^p\right)^{1/p} \]
- \(x_i\) and \(y_j\) are elements of the test and reference time series respectively
- \(v\) stands for “variable” which is for comparing multivariate series
- i.e. the \(L_p\) norm for each pair of points is summed over all variables in the multivariate series.
- \(p\) is the order of the norm used
- e.g. 1 is Manhattan distance; 2 is Euclidean
- Choice of \(p\) only matters if multivariate series are being used
Each \(\text{lcm}(i , j)\) value is an element in the \(n \times m\) matrix, \(\text{LCM}\) where \(1 \lt i \lt n\) and \(1 \lt j \lt m\)
Step Patterns
- Determines how algorithm moves across the rows of the LCM to create alignments (warpings of the time axis)
- Each pattern is a set of rules and weights
- The rules are used to create different alignments of the LCM (i.e warping of the time axis)
- The edge weights, \(m_\phi\), are used in the DTW calculation
- Patterns
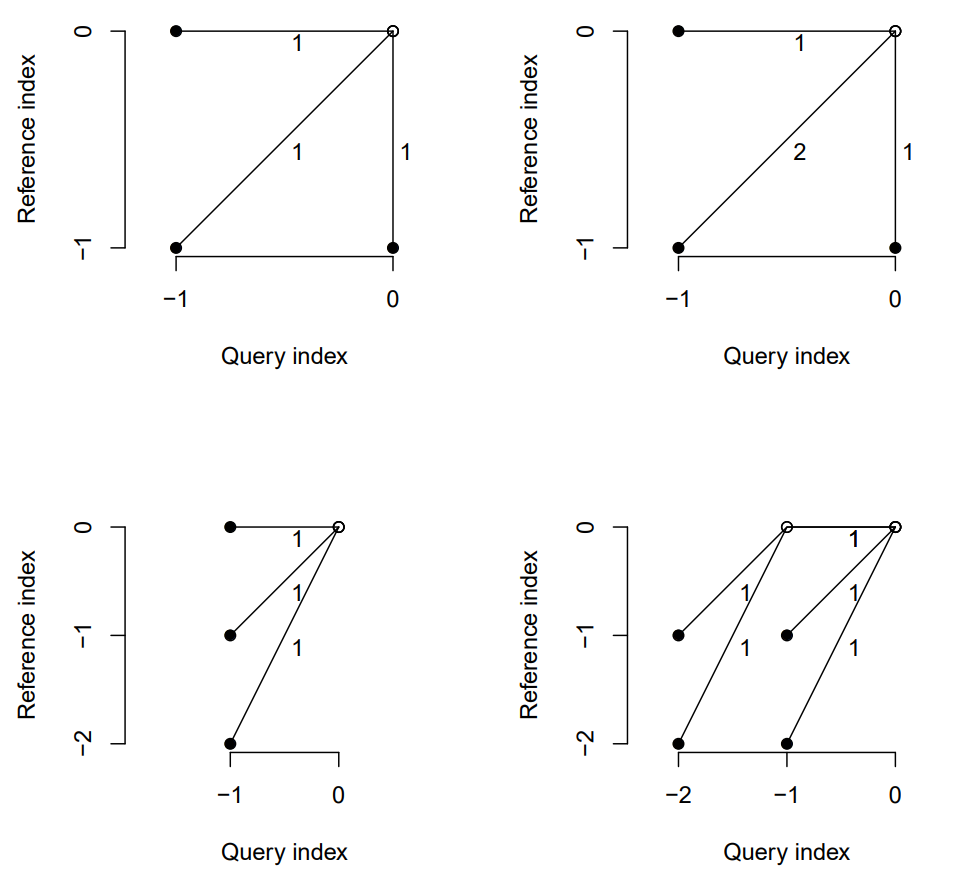
- Patterns in fig
- Top Row: symmetric1, symmetric2
- Bottom Row: asymmetric, rabinerJuangStepPattern(4, “c”, TRUE)
- i.e. Rabiner-Juang’s type IV with slope weighting
- Only some of the patterns are normalizable (i.e. \(M_\phi\) is used in the DTW equation) (normalize argument)
- Normalization may be important when
- Comparing alignments between time series of different lengths, to decide the best match (e.g., for classification)
- When performing partial matches (?)
- For
dtw_basic, doc says only supported with symmetric2 - rabinerJuangStepPattern with slope weighting types c and d are normalizable
- symmetricP* (where * is a number) are all normalizable (not shown in fig)
- Normalization may be important when
- Symmetric (i.e. dist from A to B equals the distance from B to A) only if:
- Either symmetric1 or symmetric2 step patterns are used
- Series are equal length after any constraints are used
- Some Cluster Validity Indicies (CVI) (See Clustering, Time Series >> Cluster Validity Indicies) require symmetric distance functions
- {dtwclust} author says symmetric1 most commonly used.
dtw::dtw_basicuses symmetric2 by default.
- Patterns in fig
Window Constraints
- Limits the region that the lcm calculation takes place.
- Reduces computation time but makes sense that you don’t want to compare points that are separated by to large a time interval
- Sakoe-Chiba window creates a calculation region along the diagonal (yellow blocks) of the LCM
.png)
- 1 set of lcm calculations occurs within the horizontal, rectangular block of the query series and the vertical, rectangular block of the reference series.
- Therefore, pairs are chosen in relatively small neighbors around the ith point of series 1 and the ith point of series 2, and the window size is size of the neighborhood.
- Sakoe-Chiba requires equal length series, but a “slanted band” is equivalent and works for unequal length series.
- “Slanted Band” is what’s used by dtwclust when the window constraint is used.
- Optimal window size needs to be tuned
- Can marginally speed up the DTW calculation, but they are mainly used to avoid pathological warping
- \(w\), the window size, is around half the size of the actual region covered
- \([(i, j - w), (i, j + w)]\) which has \(2w + 1\) elements
- A common w is 10% of the sample size, smaller sizes sometimes produce better results
Lower Bounds (LB)
dtw_lb- Uses a lower bound the dtw distance to speed up computation
- A considerably large dataset would be needed before the overhead of DTW becomes much larger than that of dtw_lb’s iterations
- May only be useful if one is only interested in nearest neighbors, which is usually the case in partitional clustering
- Steps
- Calculates an initial estimate of a distance matrix between two sets of time series using
lb_improved- Involves the “lower bound” calculation (Didn’t get into it)
- Uses the estimate to calculate the corresponding true DTW distance between only the nearest neighbors (row-wise minima of dist.matrix) of each series in \(x\) found in $y
- Updates distance matrix with DTW values
- Continues iteratively until no changes in the nearest neighbors occur
- Calculates an initial estimate of a distance matrix between two sets of time series using
- Only if dataset is very large will this method will be faster than
dtw_basicin the calculation of DTW - Not symmetric, no multivariate series
- Requires
- Both series to be equal length
- Window constraint defined
- Norm defined
- Value of LB (tightness of envelope around series) affected by step pattern which is set in
dtw_basicand included via … indtw_lb- Size of envelopes in general: LB_Keoghp < LB_Improvedp < DTWp
Soft DTW
sdtw- “Regularizes DTW by smoothing it” (¯\_(ツ)_/¯)
- “smoothness” controlled by gamma
- Default: 0.01
- With lower values resulting in less smoothing
- “smoothness” controlled by gamma
- Uses a gradient to efficiently calculate cluster prototypes
- Not recommended for stand-alone distance calculations
- Negative values can happen
- Symmetric and handles series of different lengths and multivariate series
Shape-based Distance (SBD)
\[ \text{SBD} (x,y) = 1-\frac{\max{\text{NCC}_c (x,y)}}{\lVert x \rVert_2 \lVert y \rVert_2} \]
SBD- Used in k-Shape Clustering
- Based on the cross-correlation with coefficient normalization (NCCc) sequence between two series
- Fast (uses FFT to calc), competitive with other distance algorithms, and supports series with different lengths
- Symmetric, no multivariate series
- In preprocessing argument, set to z-normalization
Triangular Global Alignment Kernel Distance
\[ \begin{aligned} &\text{TGAK}(x,y,\sigma,T) = \frac{\omega(i,j)\kappa(x,y)}{2-\omega(i,j)\kappa(x,y)} \\ &\begin{aligned} \text{where}\quad &\omega(i,j) = \left(1-\frac{|i-j|}{T}\right)_+ \\ &\kappa(x,y)= e^{-\phi_\sigma(x,y)} \\ &\phi_\sigma(x,y) = \frac{1}{2\sigma^2}\lVert x-y \rVert^2 + \log \left(2-e^{-\frac{\lVert x-y \rVert^2}{2\sigma^2}}\right) \end{aligned} \end{aligned} \]
GAK- “Regularizes DTW by smoothing it” (¯\_(ツ)_/¯)
- Symmetric when normalized (dist a to b = dist b to a)
- Supports multivariate series and series of different length (as long as one series isn’t half the length of the other)
- Slightly more computationally expensive than DTW (… that equation🥴)
- \(\sigma\) can defined by the user but if left as NULL, the function estimates it
- \(T\) is the triangular constraint and is similar to the window constraint in DTW but there no argument for it, so I guess it’s taken care of
- No idea what \(i\) and \(j\) refer to since x and y are also used.
- Would have to look it up in the original paper or there is a separate website and package for it
- If normalize = TRUE, then a distance is returned, can be compared with the other distance measure, and used in clustering
- If FALSE, a similarity is returned